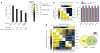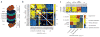A comprehensive strategy enabling high-resolution functional analysis of the yeast genome - PubMed (original) (raw)
A comprehensive strategy enabling high-resolution functional analysis of the yeast genome
David K Breslow et al. Nat Methods. 2008 Aug.
Abstract
Functional genomic studies in Saccharomyces cerevisiae have contributed enormously to our understanding of cellular processes. Their full potential, however, has been hampered by the limited availability of reagents to systematically study essential genes and the inability to quantify the small effects of most gene deletions on growth. Here we describe the construction of a library of hypomorphic alleles of essential genes and a high-throughput growth competition assay to measure fitness with unprecedented sensitivity. These tools dramatically increase the breadth and precision with which quantitative genetic analysis can be performed in yeast. We illustrate the value of these approaches by using genetic interactions to reveal new relationships between chromatin-modifying factors and to create a functional map of the proteasome. Finally, by measuring the fitness of strains in the yeast deletion library, we addressed an enigma regarding the apparent prevalence of gene dispensability and found that most genes do contribute to growth.
Figures
Figure 1
A library of hypomorphic alleles of essential yeast genes constructed using the DAmP approach. (a) Schematic of the strategy used to construct DAmP alleles. The kanamycin-resistance (KanR) cassette was inserted immediately after the open reading frame of each gene by transformation with a PCR product encoding the KanR cassette flanked at each end with homology to the targeted locus. (b) Number of DAmP diploid and haploid strains obtained. Of the 1,033 essential yeast genes, ~970 were obtained in diploid form and 842 as MATa haploids. (c) Schematic of the generation of DAmP strains and of degron-DAmP strains from existing strains with TAP tags by high-efficiency transformation with either of two universal cassettes. Deg, degron. (d) TAP-degron-DAmP alleles typically reduce protein abundance more than the DAmP allele alone as determined by western blots using an antibody to the TAP tag. To normalize for variation in loading, 3-phosphoglycerate kinase (Pgk1) was probed.
Figure 2
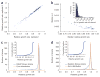
A flow cytometry–based technique for high-precision growth rate measurements. (a) Schematic of the assay. GFP-labeled mutant strains were competed with an RFP-labeled wild-type strain in 384-well format with cultures maintained by serial dilution over a 72-h time course. At each time point, samples were removed for analysis by flow cytometry to determine the ratio of GFP-positive to RFP-positive cells. (b) Wild-type and mutant cell populations can be resolved by flow cytometry. Rare events that appear to be GFP-positive and RFP-positive represent instances in which a mutant cell and a wild-type cell are misidentified by the cytometer as a single cell, and we took this into account during analysis. (c) Representative growth rate data obtained by flow cytometry. Relative growth rates spanning ~0.6 (sick) to 1.0 (wild-type growth) were observed with a high degree of linearity in the rate of mutant strain depletion over time. Dashed gray line indicates the approximate limit of sensitivity.
Figure 3
Growth rate measurements of deletion and DAmP yeast strains by the flow cytometry–based technique. (a) Correspondence between replicate measurements of relative growth rates for 344 deletion and DAmP strains. (b) Error as a function of the relative growth rate observed. For groups of 30 strains with similar growth rates, median fitness and median s.d. of triplicate measurements are shown. Inset, distribution of s.d. of triplicate measurements for all strains measured; the median s.d. was 0.0038. (c) Distribution predicted from experimental error and observed distribution of relative growth rates for 835 DAmP strains, with the inset showing the cumulative distribution of growth rates. (d) Distribution predicted from experimental error and observed distribution of relative growth rates for 4,204 deletion strains, with the inset showing the cumulative distribution of growth rates.
Figure 4
Utility of DAmP strains for identification of drug targets. (a) Sensitivity of DAmP alleles of known targets to the indicated drugs. Zones of growth inhibition surrounding drug-soaked discs were measured to determine the relative drug sensitivity of the indicated strains. (b) Distribution of growth defects for DAmP library strains grown in medium containing 0.25 μg/ml tunicamycin. Data for the strain bearing a DAmP allele of ALG7 is in red. Additional sensitive strains were also detected (dashed box; see Supplementary Data 2).
Figure 5
Highly sensitive growth rate measurements enable resolution of previously inaccessible classes of genetic interactions. (a) Schematic illustrating normalization of genetic interaction scores for an example case involving two genes, A and B. The bar graph indicates relative growth rates for the indicated strains, with the double-gene deletion growth rate shown to be equal to that expected for the case of no genetic interaction (left). For possible observed double-gene deletion growth rates, raw and normalized interaction scores are shown, along with the corresponding classes of alleviating interactions and a colorbar used for graphical representation of normalized interaction scores (right). (b) Comparison of genetic interactions observed between members of the COG complex when using fitnesses measured by flow cytometry (lower left) versus colony size (upper right; data from reference 2). Values shown for interactions calculated from flow cytometry measurements are the raw genetic interaction score (ε; black) and the normalized interaction score _(ε_NORM; red). Relative growth rates for the single-gene deletion strains are indicated in blue. (c) Growth rates measured by flow cytometry for strains with deletions of HTZ1 and/or genes encoding members of the Swr-C. Error bars indicate s.d., n ≥ 3. (d) Genetic interactions between genes encoding members of Rpd3C(S) and Rpd3C(L). The organization of these complexes is shown schematically at right (note that both complexes also contain additional members). Question marks indicate additional functions for Set2 and Rpd3C(S) suggested by genetic interaction data. Raw interaction scores (upper right) and normalized interaction scores (lower left) were calculated as in a. Note that synthetic (ε < 0) interactions are normalized using a different scale (see Supplementary Methods).
Figure 6
Functional dissection of the 26S proteasome. (a) Schematic representation of the 26S proteasome. (b) Map of raw genetic interaction scores for the 26S proteasome. Interactions between pairs of genes encoding components of the same physical subcomplex are indicated by red boxes. (c) Map of normalized interaction scores for the 26S proteasome. In the lower panel, double-mutants were grouped according to the subcomplex(es) with mutations, with the median score for each group shown (*, data for PRE9 and PRE7 were excluded when calculating the medians). Individual scores for interactions with the α ring subunits are shown at the top. Note that synthetic interactions (ε < 0) are normalized using a different procedure (see Supplementary Methods).
Similar articles
- Functional Profiling Using the Saccharomyces Genome Deletion Project Collections.
Nislow C, Wong LH, Lee AH, Giaever G. Nislow C, et al. Cold Spring Harb Protoc. 2016 Sep 1;2016(9). doi: 10.1101/pdb.prot088039. Cold Spring Harb Protoc. 2016. PMID: 27587776 - Letter to the editor. Chemical-genetic strategy for inhibiting proteasome function in Saccharomyces cerevisiae.
Howard GC, Collins GA, Tansey WP. Howard GC, et al. Yeast. 2012 Feb;29(2):93-4. doi: 10.1002/yea.1919. Epub 2011 Dec 8. Yeast. 2012. PMID: 22162073 Free PMC article. No abstract available. - Screening the yeast genome for new DNA-repair genes.
Jazayeri A, Jackson SP. Jazayeri A, et al. Genome Biol. 2002;3(4):REVIEWS1009. doi: 10.1186/gb-2002-3-4-reviews1009. Epub 2002 Mar 13. Genome Biol. 2002. PMID: 11983062 Free PMC article. Review. - CRISPR/Cas9-Mediated Genome Engineering Reveals the Contribution of the 26S Proteasome to the Extremophilic Nature of the Yeast Debaryomyces hansenii.
Spasskaya DS, Kotlov MI, Lekanov DS, Tutyaeva VV, Snezhkina AV, Kudryavtseva AV, Karpov VL, Karpov DS. Spasskaya DS, et al. ACS Synth Biol. 2021 Feb 19;10(2):297-308. doi: 10.1021/acssynbio.0c00426. Epub 2021 Jan 27. ACS Synth Biol. 2021. PMID: 33501828 - Exploring genetic interactions and networks with yeast.
Boone C, Bussey H, Andrews BJ. Boone C, et al. Nat Rev Genet. 2007 Jun;8(6):437-49. doi: 10.1038/nrg2085. Nat Rev Genet. 2007. PMID: 17510664 Review.
Cited by
- Experimental evolution of Saccharomyces cerevisiae for caffeine tolerance alters multidrug resistance and target of rapamycin signaling pathways.
Geck RC, Moresi NG, Anderson LM; yEvo Students; Brewer R, Renz TR, Taylor MB, Dunham MJ. Geck RC, et al. G3 (Bethesda). 2024 Sep 4;14(9):jkae148. doi: 10.1093/g3journal/jkae148. G3 (Bethesda). 2024. PMID: 38989875 Free PMC article. - Bidirectional promoter activity from expression cassettes can drive off-target repression of neighboring gene translation.
Powers EN, Chan C, Doron-Mandel E, Llacsahuanga Allcca L, Kim Kim J, Jovanovic M, Brar GA. Powers EN, et al. Elife. 2022 Dec 12;11:e81086. doi: 10.7554/eLife.81086. Elife. 2022. PMID: 36503721 Free PMC article. - An evolutionarily conserved bimodular domain anchors ZC3HC1 and its yeast homologue Pml39p to the nuclear basket.
Gunkel P, Iino H, Krull S, Cordes VC. Gunkel P, et al. Mol Biol Cell. 2023 May 1;34(5):ar40. doi: 10.1091/mbc.E22-09-0402. Epub 2023 Mar 1. Mol Biol Cell. 2023. PMID: 36857168 Free PMC article. - A survey of yeast genomic assays for drug and target discovery.
Smith AM, Ammar R, Nislow C, Giaever G. Smith AM, et al. Pharmacol Ther. 2010 Aug;127(2):156-64. doi: 10.1016/j.pharmthera.2010.04.012. Epub 2010 May 28. Pharmacol Ther. 2010. PMID: 20546776 Free PMC article. Review. - Phenomics approaches to understand genetic networks and gene function in yeast.
Yeung CHL, Sahin N, Andrews B. Yeung CHL, et al. Biochem Soc Trans. 2022 Apr 29;50(2):713-721. doi: 10.1042/BST20210285. Biochem Soc Trans. 2022. PMID: 35285506 Free PMC article.
References
- Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. - PubMed
- Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. - PubMed
- Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. - PubMed
- Pan X, et al. A robust toolkit for functional profiling of the yeast genome. Mol Cell. 2004;16:487–496. - PubMed
- Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–449. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous