Computational identification of ubiquitylation sites from protein sequences - PubMed (original) (raw)
Computational identification of ubiquitylation sites from protein sequences
Chun-Wei Tung et al. BMC Bioinformatics. 2008.
Abstract
Background: Ubiquitylation plays an important role in regulating protein functions. Recently, experimental methods were developed toward effective identification of ubiquitylation sites. To efficiently explore more undiscovered ubiquitylation sites, this study aims to develop an accurate sequence-based prediction method to identify promising ubiquitylation sites.
Results: We established an ubiquitylation dataset consisting of 157 ubiquitylation sites and 3676 putative non-ubiquitylation sites extracted from 105 proteins in the UbiProt database. This study first evaluates promising sequence-based features and classifiers for the prediction of ubiquitylation sites by assessing three kinds of features (amino acid identity, evolutionary information, and physicochemical property) and three classifiers (support vector machine, k-nearest neighbor, and NaïveBayes). Results show that the set of used 531 physicochemical properties and support vector machine (SVM) are the best kind of features and classifier respectively that their combination has a prediction accuracy of 72.19% using leave-one-out cross-validation.Consequently, an informative physicochemical property mining algorithm (IPMA) is proposed to select an informative subset of 531 physicochemical properties. A prediction system UbiPred was implemented by using an SVM with the feature set of 31 informative physicochemical properties selected by IPMA, which can improve the accuracy from 72.19% to 84.44%. To further analyze the informative physicochemical properties, a decision tree method C5.0 was used to acquire if-then rule-based knowledge of predicting ubiquitylation sites. UbiPred can screen promising ubiquitylation sites from putative non-ubiquitylation sites using prediction scores. By applying UbiPred, 23 promising ubiquitylation sites were identified from an independent dataset of 3424 putative non-ubiquitylation sites, which were also validated by using the obtained prediction rules.
Conclusion: We have proposed an algorithm IPMA for mining informative physicochemical properties from protein sequences to build an SVM-based prediction system UbiPred. UbiPred can predict ubiquitylation sites accompanied with a prediction score each to help biologists in identifying promising sites for experimental verification. UbiPred has been implemented as a web server and is available at http://iclab.life.nctu.edu.tw/ubipred.
Figures
Figure 1
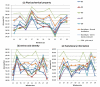
Performance comparisons among various classifiers with the three kinds of features. (a) physicochemical property, (b) amino acid identity, and (c) evolutionary information.
Figure 2
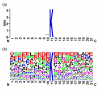
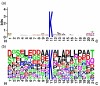
The sequence logo of the 151 positive samples with w = 21. (a) information content and (b) frequency plot.
Figure 3
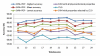
Performance comparisons between the SVM with informative physicochemical properties (SVM+IPCP) and other compared classifiers.
Figure 4
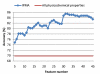
The best 10-CV accuracies of prediction using SVM with the window size 21 for various numbers of features (properties) selected by IPMA from 30 independent runs.
Figure 5
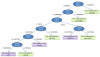
The derived decision tree by using C5.0 and the features of informative physicochemical properties for classification of ubiquitylation sites.
Figure 6
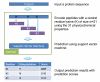
The system flow of the prediction server UbiPred.
Figure 7
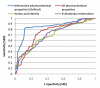
Performance comparison of SVM with various features, informative physicochemical properties (UbiPred), amino acid identity, evolutionary information, and all physicochemical properties, in terms of receiver operating characteristic curves.
Figure 8
The schema for illustrating the training data (302 samples) and the independent dataset (3424 putative non-ubiquitylation sites) using w = 21 as an example.
Figure 9
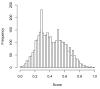
Histogram result of UbiPred using prediction scores from evaluating 3424 putative non-ubiquitylation sites in an independent dataset. The site with a score close to 1 has a high possibility to be an ubiquitylation site.
Figure 10
The sequence logo of the 23 peptides of promising ubiquitylation sites with w = 21. (a) Information content and (b) Frequency plot.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous