A unique protection signal in Cubitus interruptus prevents its complete proteasomal degradation - PubMed (original) (raw)
A unique protection signal in Cubitus interruptus prevents its complete proteasomal degradation
Yifei Wang et al. Mol Cell Biol. 2008 Sep.
Abstract
The limited proteolysis of Cubitus interruptus (Ci), the transcription factor for the developmentally and medically important Hedgehog (Hh) signaling pathway, triggers a critical switch between transcriptional repressor and activator forms. Ci repressor is formed when the C terminus of full-length Ci is degraded by the ubiquitin-proteasome pathway, an unusual reaction since the proteasome typically completely degrades its substrates. We show that several regions of Ci are required for generation of the repressor form: the zinc finger DNA binding domain, a single lysine residue (K750) near the degradation end point, and a 163-amino-acid region at the C terminus. Unlike other proteins that are partially degraded by the proteasome, dimerization is not a key feature of Ci processing. Using a pulse-chase assay in cultured Drosophila cells, we distinguish between regions required for initiation of degradation and those required for the protection of the Ci N terminus from degradation. We present a model whereby the zinc finger region and K750 together form a unique protection signal that prevents the complete degradation of Ci by the proteasome.
Figures
FIG. 1.
Kc cells are a good model for Ci proteolysis. (A) Schematic representation of Ci. ZnF, zinc fingers. The exact C terminus of Ci-75 is unknown but is estimated to lie between amino acids 650 to 700 (1). (B) Western blot of immunoprecipitates from Kc cells transfected with HACi (lane 1) and the following Ci mutants with the designated S- or T-to-A substitutions: HACi3m, the first three PKA sites (lane 2); HACi5m, all five PKA sites (lane 3); HACiNm, two GSK3 sites (lane 4); and CiCm, three CKI sites (lane 5). In this and subsequent experiments, cell extracts were immunoprecipitated with anti-HA antibody 12CA5 and Western blotted with anti-HA antibody 3F10 unless otherwise stated. (C) Western blot of immunoprecipitates from Kc cells transfected with HACi and treated with the indicated dsRNAs. The ratio of Ci-75 to Ci-155, normalized to the mock control, is shown in the box below the lane numbers. (D) Western blot of immunoprecipitates from Kc cells transfected with the indicated HACi mutants. The Ci-75-like product is approximately 30 kDa, as expected for Ci-75 lacking residues 1 to 439. The ratio of Ci-75-like protein to full-length protein, normalized to that of HACiΔN, is shown in the box below the lane numbers. Based on other experiments, CiΔN processing is reduced by ∼24% relative to wild-type Ci. (E) Western blot of immunoprecipitates from Kc cells transfected with HACiΔNΔCos2, pretreated with (+) or without (−) Cos2 dsRNA.
FIG. 2.
Multiple domains of Ci are required for its processing. (A) Western blot of immunoprecipitates from Kc cells transfected with HACi wild-type (lane 1) and the indicated HACi mutants. In this and subsequent panels, the ratio of processed form to full-length form, normalized to that of HACi, is shown in the box below the lane numbers. (B) Western blot of immunoprecipitates from Kc cells transfected with HACi wild type (lane 1) and the indicated mutants. (C) Western blot of immunoprecipitates from Kc cells transfected with HACi wild-type (lane 1) and the indicated mutants. (D) Western blot of immunoprecipitates from Kc cells transfected with HACi wild type (lane 1), HACi3m (lane 2), and HACi with deletions of the indicated zinc finger(s): ΔZn1, deletion of zinc finger 1; ΔZn2, deletion of zinc finger 2; ΔZn12, deletion of zinc fingers 1 and 2; ΔZn345, deletion of zinc fingers 3, 4, and 5. (E) Schematic drawing of the domains required for Ci proteolysis, with green indicating domains not required for Ci-75 generation, red indicating domains absolutely required for Ci-75 generation, yellow indicating domains required for the optimal processing, and orange indicating the domains together are required for optimal Ci-75 generation.
FIG. 3.
A well-folded domain, but not dimerization, is required for Ci-75 formation. (A) Schematic representation of Ci and Ci-fusion proteins. (B) Western blot of immunoprecipitates from Kc cells transfected with the indicated HACi constructs. The filled arrowhead indicates full-length proteins, and the open arrowhead indicates Ci-75 or corresponding processed products. The ratio of processed form to full-length form, normalized to that of HACi, is shown in the box below the lane numbers. (C) Western blot of immunoprecipitates (top two panels) or lysates (bottom panel) from Kc cells expressing the indicated proteins. Antibodies used for Western blotting (WB) and IP are shown to the left of the panels. The filled arrowheads indicate full-length proteins. The open arrowheads indicate Ci-75 or corresponding processed products. (D) Western blot of immunoprecipitates from Kc cells transfected with mC* and the following HACi chimeras in which the zinc fingers have been replaced by wild-type barnase or the indicated barnase mutant, with the folding stability relative to wild-type shown in parentheses: HACiZBar, wild-type barnase; HACiZBar3m, HACiZBar combined with S-to-A substitutions in three PKA sites in Ci; HACiZBarAm, barnase with three mutations (−1.66 kcal/mol); HACiZBarACm, barnase with five mutations (−2.56 kcal/mol); HACiZBarABCm, barnase with six mutations (−3.44 kcal/mol). (E) A plot of the relative folding strength of barnase mutants versus the ratio of processed product to the full-length form when the barnase mutants are substituted for the zinc finger region of Ci.
FIG. 4.
A simple, repetitive sequence is not absolutely required for Ci-75 formation. (A) Amino acid sequence of Ci from 605 to 760. The low-complexity sequences (according to the SEG algorithm with parameters 12/2.2/2.5) are highlighted in yellow, and the lysine K750 is highlighted in red. (B) Western blot of immunoprecipitates from Kc cells transfected with the indicated triple-HA-tagged proteins. The ratio of processed form to full-length form, normalized to that of HACi, is shown in the box below the lane numbers.
FIG. 5.
A single lysine residue is necessary for Ci-75 formation. (A and B) Western blot of immunoprecipitates from Kc cells expressing the indicated HACi wild-type or mutant proteins. In panel A and subsequent panels, the ratio of processed form to full-length form, normalized to that of HACi, is shown in the box below the lane numbers. Ci712Spacer refers to Ci with an insertion of a 75-amino-acid lysineless spacer after amino acid 712. (C) Schematic representation of residues 453 to 760 of wild-type Ci and the corresponding region of the Ci fusion proteins that are used in panel D. Green, red, and yellow regions are as described in the legend of Fig. 2E. Blue indicates amino acids from CG15031; pink indicates residues from mouse 4EBP1. CiCG85Ins and CiCG108Ins refer to Ci with 85 or 108 amino acids, respectively, substituted from CG15031; Ci4EBP85Ins and Ci4EBP108Ins, Ci with 85 or 108 amino acids, respectively, substituted from mouse 4EBP1. (D) Western blot of immunoprecipitates from Kc cells expressing the indicated HACi wild-type or mutant proteins. The open arrowheads indicate the Ci-75-like processed products. (E) Western blot of immunoprecipitates from Kc cells transfected with the constructs indicated above the lanes. The CiGRR fusion proteins are defined as follows: CiGRR, amino acids 615 to 760 of Ci replaced by two copies of GRR from NF-κB p105 (42); CiGRR5m, CiGRR combined with S-to-A substitutions in five PKA sites in Ci; CiGRRK5R, CiGRR combined with K782R, K792R, K795R, K822R, and K-to-R mutation of the single lysine in the GRR region. The open arrowhead indicates the Ci-75-like processed products; the filled arrowhead indicates the full-length proteins. (F) Western blots (WBs) of 9E10 IPs of cells transfected with HA-Ub and either myc-tagged wild-type Ci fragment or CiK750R fragment (amino acids 663 to 908) with (+) or without (−) Slimb dsRNA and 50 mM MG132. Western blotting was performed with anti-HA antibody (3F10) to detect conjugated HA-Ub (top panel) or anti-myc antibody to monitor the level of MycCi663-908 or MycCi663-908·K750R (bottom panel). (G) Schematic representation of residues 453 to 760 of wild-type Ci and the corresponding regions of the Ci-Ub fusion proteins that are used in panel H. Green, red and yellow indicate the same regions as described in the legend of Fig. 2E. (H) Western blot of IPs from Kc cells transfected with the indicated constructs. HACiΔZn12·4xUb, HACi4xUb with a deletion of zinc fingers 1 and 2 of Ci. The open arrowhead indicates the Ci-75-like processed product, the size of which indicates that its C terminus coincides approximately with the C-terminal end of the zinc finger region. We believe that the slower migrating products above the full-length Ci mutant band in lanes 3 and 4 are polyubiquitinated species.
FIG. 6.
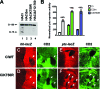
K750 is required for Ci-75 formation in vivo. (A) Western blot of lysates from embryos expressing the indicated HACi wild-type or mutant proteins. (B) Transcriptional activity of wild-type Ci, Ci3m, and CiK750R in Kc cells. Firefly and Renilla luciferase activity were measured in whole-cell extracts from Kc cells cotransfected with a ptc promoter-firefly luciferase reporter plasmid, copia-Renilla luciferase, the indicated Ci constructs, and either Act5C-Hh expressing vector (+Hh) or empty Act5C vector. Normalized luciferase activities were averaged from triplicate samples and standard errors of the means are shown. (C to F) Confocal micrographs of a part of wing imaginal discs expressing wild-type Ci (CiWT) or CiK750R transgenes throughout the disc. hh-lacZ expression (C and D) or _ptc_-lacZ expression (E and F) is detected with anti-β-galactosidase in red. Arrows point to smo mutant clones marked by the absence of CD2 staining in green. Anterior is to the left, and dorsal is to the top.
FIG. 7.
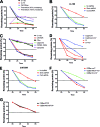
Determination of the half-lives of Ci and its mutants. (A) The half-lives of wild-type Ci-155 and Ci612Stop are determined by CHX chase in Kc cells (see Materials and Methods). Data points were plotted as the percentage of the amount of protein present at the beginning of the chase, except for Ci-75, where data points were plotted as the percentage of the amount of Ci-155 present at the beginning of the chase. The theoretical curves were calculated based on the assumption that either 30% or 100% of degradation initiation events generate Ci-75 (see Materials and Methods). (B and E) CHX chase of wild-type Ci-155 (B) and CiK750R (E) from Kc cells treated with the indicated dsRNAs. (C) CHX chase of wild-type Ci-155 and the indicated mutants. (D) CHX chase of wild-type Ci-155 and CiΔZincAll with or without coexpression of mC* as indicated. (F) CHX chase of CiZBar, CiZBarABCm (CiZBar with six mutations in barnase), and CiZBar3m with coexpression of mC*. (G) CHX chase of CiZBarStop and CiZBarABCmStop (CiZBar and CiZBarABCm, respectively, each of whose genes has a stop codon introduced after the codon for residue 612).
Similar articles
- Costal 2 interactions with Cubitus interruptus (Ci) underlying Hedgehog-regulated Ci processing.
Zhou Q, Kalderon D. Zhou Q, et al. Dev Biol. 2010 Dec 1;348(1):47-57. doi: 10.1016/j.ydbio.2010.09.004. Epub 2010 Sep 17. Dev Biol. 2010. PMID: 20850429 Free PMC article. - Drosophila hedgehog can act as a morphogen in the absence of regulated Ci processing.
Little JC, Garcia-Garcia E, Sul A, Kalderon D. Little JC, et al. Elife. 2020 Oct 21;9:e61083. doi: 10.7554/eLife.61083. Elife. 2020. PMID: 33084577 Free PMC article. - Hedgehog-induced phosphorylation by CK1 sustains the activity of Ci/Gli activator.
Shi Q, Li S, Li S, Jiang A, Chen Y, Jiang J. Shi Q, et al. Proc Natl Acad Sci U S A. 2014 Dec 30;111(52):E5651-60. doi: 10.1073/pnas.1416652111. Epub 2014 Dec 15. Proc Natl Acad Sci U S A. 2014. PMID: 25512501 Free PMC article. - Decoding Ci: from partial degradation to inhibition.
Xiong Y, Liu C, Zhao Y. Xiong Y, et al. Dev Growth Differ. 2015 Feb;57(2):98-108. doi: 10.1111/dgd.12187. Epub 2014 Dec 14. Dev Growth Differ. 2015. PMID: 25495033 Review. - Regulation of Hh/Gli signaling by dual ubiquitin pathways.
Jiang J. Jiang J. Cell Cycle. 2006 Nov 1;5(21):2457-63. doi: 10.4161/cc.5.21.3406. Epub 2006 Sep 14. Cell Cycle. 2006. PMID: 17102630 Review.
Cited by
- Costal 2 interactions with Cubitus interruptus (Ci) underlying Hedgehog-regulated Ci processing.
Zhou Q, Kalderon D. Zhou Q, et al. Dev Biol. 2010 Dec 1;348(1):47-57. doi: 10.1016/j.ydbio.2010.09.004. Epub 2010 Sep 17. Dev Biol. 2010. PMID: 20850429 Free PMC article. - Regulation of mammalian Gli proteins by Costal 2 and PKA in Drosophila reveals Hedgehog pathway conservation.
Marks SA, Kalderon D. Marks SA, et al. Development. 2011 Jun;138(12):2533-42. doi: 10.1242/dev.063479. Development. 2011. PMID: 21610030 Free PMC article. - Ligand-independent activation of the Hedgehog pathway displays non-cell autonomous proliferation during eye development in Drosophila.
Christiansen AE, Ding T, Bergmann A. Christiansen AE, et al. Mech Dev. 2012 Jul;129(5-8):98-108. doi: 10.1016/j.mod.2012.05.009. Epub 2012 Jun 5. Mech Dev. 2012. PMID: 22677792 Free PMC article. - Drosophila hedgehog can act as a morphogen in the absence of regulated Ci processing.
Little JC, Garcia-Garcia E, Sul A, Kalderon D. Little JC, et al. Elife. 2020 Oct 21;9:e61083. doi: 10.7554/eLife.61083. Elife. 2020. PMID: 33084577 Free PMC article. - Proteostasis in the Hedgehog signaling pathway.
Liu A. Liu A. Semin Cell Dev Biol. 2019 Sep;93:153-163. doi: 10.1016/j.semcdb.2018.10.009. Epub 2018 Nov 14. Semin Cell Dev Biol. 2019. PMID: 31429406 Free PMC article. Review.
References
- Aza-Blanc, P., F. A. Ramirez-Weber, M. P. Laget, C. Schwartz, and T. B. Kornberg. 1997. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 891043-1053. - PubMed
- Butt, T. R., M. I. Khan, J. Marsh, D. J. Ecker, and S. T. Crooke. 1988. Ubiquitin-metallothionein fusion protein expression in yeast. A genetic approach for analysis of ubiquitin functions. J. Biol. Chem. 26316364-16371. - PubMed
- Chen, C. H., D. P. von Kessler, W. Park, B. Wang, Y. Ma, and P. A. Beachy. 1999. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell 98305-316. - PubMed
- Dai, P., H. Akimaru, and S. Ishii. 2003. A hedgehog-responsive region in the Drosophila wing disc is defined by debra-mediated ubiquitination and lysosomal degradation of Ci. Dev. Cell 4917-928. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases