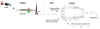XCMS2: processing tandem mass spectrometry data for metabolite identification and structural characterization - PubMed (original) (raw)
. 2008 Aug 15;80(16):6382-9.
doi: 10.1021/ac800795f. Epub 2008 Jul 16.
Affiliations
- PMID: 18627180
- PMCID: PMC2728033
- DOI: 10.1021/ac800795f
XCMS2: processing tandem mass spectrometry data for metabolite identification and structural characterization
H P Benton et al. Anal Chem. 2008.
Abstract
Mass spectrometry based metabolomics represents a new area for bioinformatics technology development. While the computational tools currently available such as XCMS statistically assess and rank LC-MS features, they do not provide information about their structural identity. XCMS(2) is an open source software package which has been developed to automatically search tandem mass spectrometry (MS/MS) data against high quality experimental MS/MS data from known metabolites contained in a reference library (METLIN). Scoring of hits is based on a "shared peak count" method that identifies masses of fragment ions shared between the analytical and reference MS/MS spectra. Another functional component of XCMS(2) is the capability of providing structural information for unknown metabolites, which are not in the METLIN database. This "similarity search" algorithm has been developed to detect possible structural motifs in the unknown metabolite which may produce characteristic fragment ions and neutral losses to related reference compounds contained in METLIN, even if the precursor masses are not the same.
Figures
Figure 1
The general workflow of XCMS and XCMS2 employing the “sniper” approach: a single feature is found with statistical confidence and selected for MS/MS. The data from the MS/MS is then put through XCMS2 and the feature is structurally identified using the METLIN database.
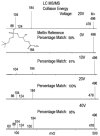
Figure 2
The comparison of the different matches at the reference collision energy. As the voltage increases, more low mass fragment ions are generated. Using a collision energy that is close to the experimentally used value gives a high matching score.
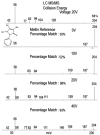
Figure 3
The difference between the collision energies is seen again. However, the figure also shows how high collision energy spectra can match lower collision energy spectra due to signal-to-noise levels of low abundance peaks.
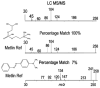
Figure 4
A METLIN search using accurate mass only can yield multiple hits. However, searching with MS/MS data, glycerophosphocholine can be unambiguously identified.
Figure 5
Similarity search. The first spectrum is the MS/MS data collected from an experiment of 1-palitylophosphocholine. The spectra below show similarity to the unknown.
Similar articles
- Autonomous METLIN-Guided In-source Fragment Annotation for Untargeted Metabolomics.
Domingo-Almenara X, Montenegro-Burke JR, Guijas C, Majumder EL, Benton HP, Siuzdak G. Domingo-Almenara X, et al. Anal Chem. 2019 Mar 5;91(5):3246-3253. doi: 10.1021/acs.analchem.8b03126. Epub 2019 Feb 11. Anal Chem. 2019. PMID: 30681830 Free PMC article. - METLIN: a metabolite mass spectral database.
Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. Smith CA, et al. Ther Drug Monit. 2005 Dec;27(6):747-51. doi: 10.1097/01.ftd.0000179845.53213.39. Ther Drug Monit. 2005. PMID: 16404815 - An efficient algorithm for the blocked pattern matching problem.
Deng F, Wang L, Liu X. Deng F, et al. Bioinformatics. 2015 Feb 15;31(4):532-8. doi: 10.1093/bioinformatics/btu678. Epub 2014 Oct 15. Bioinformatics. 2015. PMID: 25322837 - METLIN: A Tandem Mass Spectral Library of Standards.
Montenegro-Burke JR, Guijas C, Siuzdak G. Montenegro-Burke JR, et al. Methods Mol Biol. 2020;2104:149-163. doi: 10.1007/978-1-0716-0239-3_9. Methods Mol Biol. 2020. PMID: 31953817 Free PMC article. Review. - Identification of small molecules using accurate mass MS/MS search.
Kind T, Tsugawa H, Cajka T, Ma Y, Lai Z, Mehta SS, Wohlgemuth G, Barupal DK, Showalter MR, Arita M, Fiehn O. Kind T, et al. Mass Spectrom Rev. 2018 Jul;37(4):513-532. doi: 10.1002/mas.21535. Epub 2017 Apr 24. Mass Spectrom Rev. 2018. PMID: 28436590 Free PMC article. Review.
Cited by
- A Perspective on the Confident Comparison of Glycoprotein Site-Specific Glycosylation in Sample Cohorts.
Klein JA, Zaia J. Klein JA, et al. Biochemistry. 2020 Sep 1;59(34):3089-3097. doi: 10.1021/acs.biochem.9b00730. Epub 2019 Dec 31. Biochemistry. 2020. PMID: 31833756 Free PMC article. Review. - Trace, Machine Learning of Signal Images for Trace-Sensitive Mass Spectrometry: A Case Study from Single-Cell Metabolomics.
Liu Z, Portero EP, Jian Y, Zhao Y, Onjiko RM, Zeng C, Nemes P. Liu Z, et al. Anal Chem. 2019 May 7;91(9):5768-5776. doi: 10.1021/acs.analchem.8b05985. Epub 2019 Apr 15. Anal Chem. 2019. PMID: 30929422 Free PMC article. - Mapping the metabolism of five amino acids in bloodstream form Trypanosoma brucei using U-13C-labelled substrates and LC-MS.
Johnston K, Kim DH, Kerkhoven EJ, Burchmore R, Barrett MP, Achcar F. Johnston K, et al. Biosci Rep. 2019 May 17;39(5):BSR20181601. doi: 10.1042/BSR20181601. Print 2019 May 31. Biosci Rep. 2019. PMID: 31028136 Free PMC article. - Bioinformatics: the next frontier of metabolomics.
Johnson CH, Ivanisevic J, Benton HP, Siuzdak G. Johnson CH, et al. Anal Chem. 2015 Jan 6;87(1):147-56. doi: 10.1021/ac5040693. Epub 2014 Nov 20. Anal Chem. 2015. PMID: 25389922 Free PMC article. Review. No abstract available. - Simultaneous Metabolite, Protein, Lipid Extraction (SIMPLEX): A Combinatorial Multimolecular Omics Approach for Systems Biology.
Coman C, Solari FA, Hentschel A, Sickmann A, Zahedi RP, Ahrends R. Coman C, et al. Mol Cell Proteomics. 2016 Apr;15(4):1453-66. doi: 10.1074/mcp.M115.053702. Epub 2016 Jan 26. Mol Cell Proteomics. 2016. PMID: 26814187 Free PMC article.
References
- Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. Anal Chem. 2006;78:779–787. - PubMed
- Want EJ, Nordstrom A, Morita H, Siuzdak G. J Proteome Res. 2007;6:459–468. - PubMed
- Taylor CF, Paton NW, Garwood KL, Kirby PD, Stead DA, Yin Z, Deutsch EW, Selway L, Walker J, Riba-Garcia I, Mohammed S, Deery MJ, Howard JA, Dunkley T, Aebersold R, Kell DB, Lilley KS, Roepstorff P, Yates JR, 3rd, Brass A, Brown AJ, Cash P, Gaskell SJ, Hubbard SJ, Oliver SG. Nat Biotechnol. 2003;21:247–254. - PubMed
- Broeckling CD, Reddy IR, Duran AL, Zhao X, Sumner LW. Anal Chem. 2006;78:4334–4341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R24 EY017540-02/EY/NEI NIH HHS/United States
- P30 MH062261/MH/NIMH NIH HHS/United States
- P30 MH062261-05S1/MH/NIMH NIH HHS/United States
- R24EY017540/EY/NEI NIH HHS/United States
- R24 EY017540/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources