Viral detection by electron microscopy: past, present and future - PubMed (original) (raw)
Viral detection by electron microscopy: past, present and future
Philippe Roingeard. Biol Cell. 2008 Aug.
Abstract
Viruses are very small and most of them can be seen only by TEM (transmission electron microscopy). TEM has therefore made a major contribution to virology, including the discovery of many viruses, the diagnosis of various viral infections and fundamental investigations of virus-host cell interactions. However, TEM has gradually been replaced by more sensitive methods, such as the PCR. In research, new imaging techniques for fluorescence light microscopy have supplanted TEM, making it possible to study live cells and dynamic interactions between viruses and the cellular machinery. Nevertheless, TEM remains essential for certain aspects of virology. It is very useful for the initial identification of unknown viral agents in particular outbreaks, and is recommended by regulatory agencies for investigation of the viral safety of biological products and/or the cells used to produce them. In research, only TEM has a resolution sufficiently high for discrimination between aggregated viral proteins and structured viral particles. Recent examples of different viral assembly models illustrate the value of TEM for improving our understanding of virus-cell interactions.
Figures
Figure 1
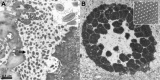
Diagnosis of virus infections by examination of ultrathin sections of human tissues or cells (A) Parapoxvirus (Orf virus) infection on a human skin biopsy specimen. Multiple oval viral particles (arrow), comprising a dense core surrounded by an envelope (inset, high magnification), are observed in an infected cell. The Orf virus is a parapoxivirus that causes a common skin disease of sheep and goats, and is occasionally transmitted to human. (B) Polyomavirus (BK virus) infection in cells pelleted from a urine sample taken from an organ‐transplant patient. The presence of a large number of viral particles leads to their arrangement into a crystal‐like structure (inset, high magnification).
Figure 2
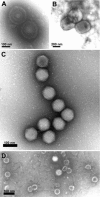
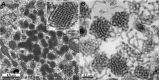
Direct negative staining of virus in fluid recovered from human skin vesicles (A and B) or from stool samples (C and D) (A and B) Rapid morphological diagnosis and differential diagnosis from a herpesvirus (Varicella, A) and a parapoxvirus (Orf virus, B). The penetration of the negative stain into the herpesvirus particle may reveal the presence of the viral capsid within the envelope. (C and D) Negative staining of viruses involved in gastroenteritis reveals the surface detail of the subunit arrangement of the adenovirus core particle (C), showing its icosahedral form clearly, whereas the rotavirus displays its typical ‘wheel‐like’ appearance (D).
Figure 3
Detection of retroviruses in rodent hybridoma cells used for the production of biological products Ultrathin sections of cells of different origins may show intracisternal A‐type retroviral particles (A) or C‐type retroviral particles budding at the cell surface (B). The C‐type particles released by the cells can be detected by negative staining in the cell supernatant (B, inset).
Figure 4
Ultrastructural changes associated with viral replication or viral factories (A) The Semliki forest virus, an alphavirus, induces the formation of a cytopathic vacuole (CPV), surrounded by the ER. Numerous viral replication complexes (arrow) are anchored in the internal membrane of the CPV. (B) The non‐structural proteins of HCV, a flavivirus, induce the formation of a membranous web in the perinuclear area.
Figure 5
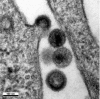
Budding of HIV The viral particle at the top shows virus formation with distortion of a cellular membrane away from the cytoplasm. The budding particle and the particle at the bottom are immature viral particles, whereas the two particles in the centre are mature and have a truncated cone‐shaped core. Thus maturation of the core by the viral protease occurs shortly after the release of the particle from the host cell membrane.
Figure 6
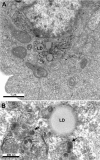
Budding of HCV Ultrastructural analysis of cells producing the HCV core protein shows that this protein self‐assembles into HCV‐like particles (arrows) at convoluted and electron‐dense ER membranes surrounding the lipid droplets (LD) present in the perinuclear area.
Figure 7
Hepatitis B subviral envelope particle morphogenesis and intracellular trafficking Ultrastructural analysis of cells producing HBV major envelope protein shows that this protein self‐assembles in the ER into filaments packed into crystal‐like structures (A, see also a high magnification of these packed filaments in the inset). These filaments are transported to the ERGIC, where they are unpacked and relaxed (B).
Similar articles
- [Virus detection by electron microscopy: past, present and future].
Roingeard P. Roingeard P. Virologie (Montrouge). 2009 Oct 1;13(5):249-258. doi: 10.1684/13-5.2011.249-258-article-1. Virologie (Montrouge). 2009. PMID: 36151618 French. - Simultaneous detection of multiplex-amplified human immunodeficiency virus type 1 RNA, hepatitis C virus RNA, and hepatitis B virus DNA using a flow cytometer microsphere-based hybridization assay.
Defoort JP, Martin M, Casano B, Prato S, Camilla C, Fert V. Defoort JP, et al. J Clin Microbiol. 2000 Mar;38(3):1066-71. doi: 10.1128/JCM.38.3.1066-1071.2000. J Clin Microbiol. 2000. PMID: 10698998 Free PMC article. - Visualizing the Essential Role of Complete Virion Assembly Machinery in Efficient Hepatitis C Virus Cell-to-Cell Transmission by a Viral Infection-Activated Split-Intein-Mediated Reporter System.
Zhao F, Zhao T, Deng L, Lv D, Zhang X, Pan X, Xu J, Long G. Zhao F, et al. J Virol. 2017 Jan 3;91(2):e01720-16. doi: 10.1128/JVI.01720-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27852847 Free PMC article. - [Molecular diagnosis of hepatitis B virus and hepatitis C virus infection].
Nomura F, Itoga S. Nomura F, et al. Rinsho Byori. 2002 Nov;Suppl 123:31-6. Rinsho Byori. 2002. PMID: 12652787 Review. Japanese.
Cited by
- Combining array tomography with electron tomography provides insights into leakiness of the blood-brain barrier in mouse cortex.
Kislinger G, Fabig G, Wehn A, Rodriguez L, Jiang H, Niemann C, Klymchenko AS, Plesnila N, Misgeld T, Müller-Reichert T, Khalin I, Schifferer M. Kislinger G, et al. Elife. 2024 Aug 5;12:RP90565. doi: 10.7554/eLife.90565. Elife. 2024. PMID: 39102289 Free PMC article. - Polysaccharides and Their Derivatives as Potential Antiviral Molecules.
Claus-Desbonnet H, Nikly E, Nalbantova V, Karcheva-Bahchevanska D, Ivanova S, Pierre G, Benbassat N, Katsarov P, Michaud P, Lukova P, Delattre C. Claus-Desbonnet H, et al. Viruses. 2022 Feb 18;14(2):426. doi: 10.3390/v14020426. Viruses. 2022. PMID: 35216019 Free PMC article. Review. - Ultrastructural and Immunohistochemical Diagnosis of a Neonatal Herpes Simplex Virus Infection Presenting as Fulminant Hepatitis: A Case Report.
Papa V, Salfi NCM, Costa R, Bettocchi I, Ricci E, Cordelli DM, Locatelli F, Caramelli F, Cenacchi G. Papa V, et al. Adv Exp Med Biol. 2022;1369:93-100. doi: 10.1007/5584_2021_659. Adv Exp Med Biol. 2022. PMID: 34302289 - Putative morphology of Neoehrlichia mikurensis in salivary glands of Ixodes ricinus.
Ondruš J, Kulich P, Sychra O, Široký P. Ondruš J, et al. Sci Rep. 2020 Sep 29;10(1):15987. doi: 10.1038/s41598-020-72953-0. Sci Rep. 2020. PMID: 32994495 Free PMC article. - MPXV: Update on Morphological and Morphogenesis Aspects Through Transmission and Scanning Electron Microscopies and 3D Reconstruction.
Barreto-Vieira DF, Miranda MD, da Silva MAN, de Almeida AS, de Almeida ALT, Bandeira DM, Ferreira VNS, Rosa AS, Girard-Dias W, Archanjo BS, Barth OM. Barreto-Vieira DF, et al. J Med Virol. 2025 Jan;97(1):e70180. doi: 10.1002/jmv.70180. J Med Virol. 2025. PMID: 39825732 Free PMC article.
References
- Appleton H. Higgins P.G. Viruses and gastroenteritis in infants. Lancet. 1975;i:1297. - PubMed
- Biel S.S. Madeley D. Diagnostic virology—the need for electron microscopy: a discussion paper J. Clin. Virol. 2001. 22 1–9 - PubMed
- Bishop R.F. Davidson G.P. Holmes I.H. Ruck B.J. Virus particles in epithelial cells of duodenal mucosa from children with acute non‐bacterial gastroenteritis Lancet 1973. ii 1281–1283 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous