Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients - PubMed (original) (raw)
Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients
Feng Wang-Johanning et al. Cancer Res. 2008.
Abstract
Recent evidence indicates that human cancer cells reactivate the expression of latent human endogenous retroviral (HERV) proteins. However, the extent to which cancer patients mount de novo immune responses against expressed HERV elements is unclear. In this study, we determined the extent of HERV-K env expression in human breast cancer (BC) and whether both humoral and cell-mediated immunity against HERV-K can be found in BC patients. We found HERV-K env protein expression in 88% of BC (n = 119) but not in normal breast (n = 76) tissues. ELISA screening assays detected significant titers of anti-HERV-K env IgG in a large proportion of BC patients. T-cell responses against HERV-K were also detected in peripheral blood mononuclear cells (PBMC) from BC patients stimulated with autologous dendritic cells pulsed with HERV-K env SU antigens. These responses included induction of T-cell proliferation (P = 0.0043), IFN-gamma production measured by enzyme-linked immunospot (P < 0.0001), and multiplex cytokine secretion (P = 0.0033). Multiplex cytokine analysis found a T-helper 1 cytokine response, including interleukin (IL)-2 (P = 0.0109), IL-6 (P = 0.0396), IL-8 (P = 0.0169), and IP-10 (P = 0.0045) secretion during in vitro stimulation of BC PBMC with HERV-K antigen. We also found HERV-K-specific CTLs that were capable of lysing target cells expressing HERV-K env protein in BC patients but not in normal female controls without cancer. These findings suggest that retroviral gene products are capable of acting as tumor-associated antigens activating both T-cell and B-cell responses in BC patients.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures
Figure 1
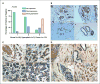
The expression profiles of HERV-K env protein in breast tissue arrays. A, summary of HERV-K env protein expression in arrays of 182 breast tissue samples, including normal breast tissues from healthy patients without cancer (n = 56), benign breast tissues from breast hyperplasia (n = 7), and BC tissues (n = 119). B, panels 1 to 4, representative samples from multiple breast tissue array slides stained with 6H5. The expression of HERV-K env protein was detected in two BC biopsies (panels 1 and 2) but not in normal mammary tissues (panels 3 and 4). C, detection of HERV-K env protein expression in one 48-year-old female diagnosed with infiltrating mammary carcinoma. Positive-staining tumor epithelial cells (brown color indicated with red arrow) were detected in DCIS (panel 1; magnification, ×40), IDC (panel 2), and metastases to the lymph node (panel 4). Positive-staining was greatly reduced (panel 3; black arrows indicate negative staining) in adjacent uninvolved epithelial cells. Red arrow, however, a few tumor cells were positively stained. D, detection of HERV-K env protein expression in one 59-year-old female diagnosed with IDC. All of the panels in this figure have regions of IDC, DCIS, uninvolved epithelial cells, and normal cells. Positive staining (brown color indicated with red arrows) was detected in tumor epithelial cells from DCIS (panel 1; magnification, ×20) and IDC (panels 1 to 4) but not in normal or uninvolved epithelial cells (negative staining indicated with black arrows in panels 2 and 3). More than 91% (31 of 34 cases) of biopsies containing >70% tumor cells had intermediate or strong expression of HERV-K env protein.
Figure 2
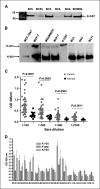
Detection of HERV-K env SU protein and anti–HERV-K IgG in patient sera. A, HERV-K env SU fusion protein K-GST was used to detect anti–HERV-K IgG in patient sera by immunoblot analysis. Antibodies against HERV-K env protein were higher in sera obtained from some BC patients (BC3, BC2, and BC9824) than others (BC53 and BC4), as shown by immunoblot using a 6H5 mAb. B, HERV-K env proteins immunoprecipitated from MCF-7 and MDA-MB-231 cells with mAb 6H5 were used to detect anti–HERV-K antibody in patient sera by immunoblot. Left blot, 6H5 mAb detected HERV-K env protein and SU protein in MCF-7 and the weakly expressing MCF10AT precancerous cell line. Full-length K-env (67 kDa) and the K-SU (30 kDa) proteins were recognized by the 6H5 mAb. The amount of IgG against HERV-K env SU protein was greater than the amount of IgG against full-length env protein precipitated from MDA-MB-231 and MCF-7 cells (middle blot) in the serum of patient BC2. K-GST fusion protein, which served as a positive control, was also detected by BC2 serum. Right blot, furthermore, HERV-K env SU proteins from BC3 and BC11 sera were immunoprecipitated by 6H5. Serum obtained from an ovarian cancer patient (OV4) was used as a control. C, titration of antibodies against HERV-K env SU protein in sera from BC patients or normal female donors using ELISA. Anti–HERV-K SU antibody titers were significantly higher in BC patients (n = 39) than in normal donors (n = 20, unpaired Student’s t test). D, proteins from type 1 HERV-K surface (K1-SU; without the 292-bp insert), gag (K-gag), and type 2 HERV-K surface (K2-SU; with the 292-bp insert) were used to detect their respective antibodies in the sera (1:200 dilution) by ELISA. Specific IgGs were detected in BC patients BC31, BC32, BC33, BC34, BC39, and BC40.
Figure 3
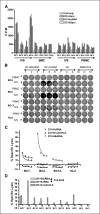
Detection of HERV-K–specific T-cell responses in BC patient PBMC. A, T-cell proliferation was compared in freshly isolated (ex vivo) PBMC versus IVS cells pulsed with HERV-K cRNA from four BC patients and four normal donors. Higher T-cell proliferation was observed only in IVS cells obtained from BC patients (BC1 and BC4). There was no significant difference in proliferation between IVS performed with DC pulsed with HERV-K cRNA or HERV-K protein. B, IFN-γ ELISPOT was used to determine HERV-K–specific T-cell response. A representative ELISPOT result is shown. More spots were detected in IVS cells (1 × 105 loaded) from BC patients (BC4, BC5, and BC17) than a normal donor (NL6) stimulated with HERV-K env cRNA. C, specific lysis by CTL was measured in PBMC after IVS from HLA-A2+ BC in comparison with normal donor PBMC using MDA-MB-231 (HLA-A2.1+) as target cells. IVS cells obtained from the three BC patients (BC7, BC2, and BC12), but not a normal subject (NL2), had increased killing of MDA-MB-231 cells transfected with HERV-K cRNA than MDA-MB-231 cells transfected with control E6 cRNA. D, HERV-K–specific T-cell killing was observed only in PBMC from patients with BC after IVS, as seen by the higher lysis of MDA-MB-231 cells transfected with HERV-K cRNA than MDA-MB-231 cells transfected with control E6 cRNA (P = 0.0255, paired Student’s t test). CTL lysis was blocked by anti–HLA-A2 antibody (231+_K_+ anti-HLA). The percentages of CTL-specific lysis of MDA-MB-231 cells expressing various antigens were compared, and lysis of only target BC cells expressing HERV-K was observed in PBMC obtained from BC patients after IVS.
Similar articles
- Cytotoxicity of human endogenous retrovirus K-specific T cells toward autologous ovarian cancer cells.
Rycaj K, Plummer JB, Yin B, Li M, Garza J, Radvanyi L, Ramondetta LM, Lin K, Johanning GL, Tang DG, Wang-Johanning F. Rycaj K, et al. Clin Cancer Res. 2015 Jan 15;21(2):471-83. doi: 10.1158/1078-0432.CCR-14-0388. Epub 2014 Nov 4. Clin Cancer Res. 2015. PMID: 25370465 - Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors.
Wang-Johanning F, Rycaj K, Plummer JB, Li M, Yin B, Frerich K, Garza JG, Shen J, Lin K, Yan P, Glynn SA, Dorsey TH, Hunt KK, Ambs S, Johanning GL. Wang-Johanning F, et al. J Natl Cancer Inst. 2012 Feb 8;104(3):189-210. doi: 10.1093/jnci/djr540. Epub 2012 Jan 12. J Natl Cancer Inst. 2012. PMID: 22247020 Free PMC article. - Human endogenous retroviruses and cancer prevention: evidence and prospects.
Cegolon L, Salata C, Weiderpass E, Vineis P, Palù G, Mastrangelo G. Cegolon L, et al. BMC Cancer. 2013 Jan 3;13:4. doi: 10.1186/1471-2407-13-4. BMC Cancer. 2013. PMID: 23282240 Free PMC article. - Molecular biology of type A endogenous retrovirus.
Ono M. Ono M. Kitasato Arch Exp Med. 1990 Sep;63(2-3):77-90. Kitasato Arch Exp Med. 1990. PMID: 1710682 Review. - Anti-HERV-K Drugs and Vaccines, Possible Therapies against Tumors.
Hosseiniporgham S, Sechi LA. Hosseiniporgham S, et al. Vaccines (Basel). 2023 Mar 28;11(4):751. doi: 10.3390/vaccines11040751. Vaccines (Basel). 2023. PMID: 37112663 Free PMC article. Review.
Cited by
- Evolution of Repetitive Elements, Their Roles in Homeostasis and Human Disease, and Potential Therapeutic Applications.
Snowbarger J, Koganti P, Spruck C. Snowbarger J, et al. Biomolecules. 2024 Oct 2;14(10):1250. doi: 10.3390/biom14101250. Biomolecules. 2024. PMID: 39456183 Free PMC article. Review. - A T cell receptor specific for an HLA-A*03:01-restricted epitope in the endogenous retrovirus ERV-K-Env exhibits limited recognition of its cognate epitope.
Grundy EE, Shaw LC, Wang L, Lee AV, Argueta JC, Powell DJ Jr, Ostrowski M, Jones RB, Cruz CRY, Gordish-Dressman H, Chappell NP, Bollard CM, Chiappinelli KB. Grundy EE, et al. Mob DNA. 2024 Oct 9;15(1):19. doi: 10.1186/s13100-024-00333-w. Mob DNA. 2024. PMID: 39385229 Free PMC article. - Epigenetic therapy potentiates transposable element transcription to create tumor-enriched antigens in glioblastoma cells.
Jang HJ, Shah NM, Maeng JH, Liang Y, Basri NL, Ge J, Qu X, Mahlokozera T, Tzeng SC, Williams RB, Moore MJ, Annamalai D, Chen JY, Lee HJ, DeSouza PA, Li D, Xing X, Kim AH, Wang T. Jang HJ, et al. Nat Genet. 2024 Sep;56(9):1903-1913. doi: 10.1038/s41588-024-01880-x. Epub 2024 Sep 2. Nat Genet. 2024. PMID: 39223316 - Limited Immunogenicity of an HLA-A*03:01-restricted Epitope of Erv-k-env in Non-hiv-1 Settings: Implications for Adoptive Cell Therapy in Cancer.
Grundy EE, Shaw LC, Wang L, Powell DJ, Ostrowski M, Jones RB, Cruz CRY, Gordish-Dressman H, Bollard CM, Chiappinelli KB. Grundy EE, et al. Res Sq [Preprint]. 2024 May 30:rs.3.rs-4432372. doi: 10.21203/rs.3.rs-4432372/v1. Res Sq. 2024. PMID: 38854052 Free PMC article. Updated. Preprint. - Endogenous retroviral solo-LTRs in human genome.
Chen M, Huang X, Wang C, Wang S, Jia L, Li L. Chen M, et al. Front Genet. 2024 Mar 28;15:1358078. doi: 10.3389/fgene.2024.1358078. eCollection 2024. Front Genet. 2024. PMID: 38606358 Free PMC article. Review.
References
- Fetsch PA, Marincola FM, Abati A. The new melanoma markers: MART-1 and Melan-A (the NIH experience) Am J Surg Pathol. 1999;23:607–10. - PubMed
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical