The protein information and property explorer: an easy-to-use, rich-client web application for the management and functional analysis of proteomic data - PubMed (original) (raw)
The protein information and property explorer: an easy-to-use, rich-client web application for the management and functional analysis of proteomic data
H Ramos et al. Bioinformatics. 2008.
Abstract
Motivation: Mass spectrometry experiments in the field of proteomics produce lists containing tens to thousands of identified proteins. With the protein information and property explorer (PIPE), the biologist can acquire functional annotations for these proteins and explore the enrichment of the list, or fraction thereof, with respect to functional classes. These protein lists may be saved for access at a later time or different location. The PIPE is interoperable with the Firegoose and the Gaggle, permitting wide-ranging data exploration and analysis. The PIPE is a rich-client web application which uses AJAX capabilities provided by the Google Web Toolkit, and server-side data storage using Hibernate.
Availability: http://pipe.systemsbiology.net.
Figures
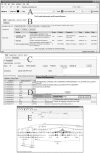
Fig. 1.
(A) Through the Firegoose, the PIPE and the Gaggle interchange data at the click of the ‘Broadcast’ button. (B) Logging into the PIPE produces a view summarizing all previously entered datasets. (C) Opening a dataset produces a view of the data and a menu bar containing operations which can be performed on the data. Here, we have performed an ID Mapping operation from IPI numbers to Entrez Gene ID, gene symbol and description. (D) When no mapping is found for a protein identifier, clicking on the ‘na’ value gives the user the option to lookup the protein sequence or enter the missing value. (E) The results of a GO enrichment operation are presented in a Gaggled Cytoscape window (Shannon et al., 2003, 2006).
Similar articles
- The Protein Information and Property Explorer 2: gaggle-like exploration of biological proteomic data within one webpage.
Ramos H, Shannon P, Brusniak MY, Kusebauch U, Moritz RL, Aebersold R. Ramos H, et al. Proteomics. 2011 Jan;11(1):154-8. doi: 10.1002/pmic.201000459. Epub 2010 Dec 6. Proteomics. 2011. PMID: 21182202 Free PMC article. - The Gaggle: an open-source software system for integrating bioinformatics software and data sources.
Shannon PT, Reiss DJ, Bonneau R, Baliga NS. Shannon PT, et al. BMC Bioinformatics. 2006 Mar 28;7:176. doi: 10.1186/1471-2105-7-176. BMC Bioinformatics. 2006. PMID: 16569235 Free PMC article. - Qupe--a Rich Internet Application to take a step forward in the analysis of mass spectrometry-based quantitative proteomics experiments.
Albaum SP, Neuweger H, Fränzel B, Lange S, Mertens D, Trötschel C, Wolters D, Kalinowski J, Nattkemper TW, Goesmann A. Albaum SP, et al. Bioinformatics. 2009 Dec 1;25(23):3128-34. doi: 10.1093/bioinformatics/btp568. Epub 2009 Oct 6. Bioinformatics. 2009. PMID: 19808875 - Elective affinities--bioinformatic analysis of proteomic mass spectrometry data.
Li X, Pizarro A, Grosser T. Li X, et al. Arch Physiol Biochem. 2009 Dec;115(5):311-9. doi: 10.3109/13813450903390039. Arch Physiol Biochem. 2009. PMID: 19911947 Review. - Recent developments in public proteomic MS repositories and pipelines.
Mead JA, Bianco L, Bessant C. Mead JA, et al. Proteomics. 2009 Feb;9(4):861-81. doi: 10.1002/pmic.200800553. Proteomics. 2009. PMID: 19212957 Review.
Cited by
- ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks.
Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. Bindea G, et al. Bioinformatics. 2009 Apr 15;25(8):1091-3. doi: 10.1093/bioinformatics/btp101. Epub 2009 Feb 23. Bioinformatics. 2009. PMID: 19237447 Free PMC article. - SIDEKICK: Genomic data driven analysis and decision-making framework.
Doderer MS, Yoon K, Robbins KA. Doderer MS, et al. BMC Bioinformatics. 2010 Dec 30;11:611. doi: 10.1186/1471-2105-11-611. BMC Bioinformatics. 2010. PMID: 21192813 Free PMC article. - Differential Plasma Glycoproteome of p19 Skin Cancer Mouse Model Using the Corra Label-Free LC-MS Proteomics Platform.
Letarte S, Brusniak MY, Campbell D, Eddes J, Kemp CJ, Lau H, Mueller L, Schmidt A, Shannon P, Kelly-Spratt KS, Vitek O, Zhang H, Aebersold R, Watts JD. Letarte S, et al. Clin Proteomics. 2008 Dec 1;4(3-4):105. doi: 10.1007/s12014-008-9018-8. Clin Proteomics. 2008. PMID: 20157627 Free PMC article. - Integrated phosphoproteomics analysis of a signaling network governing nutrient response and peroxisome induction.
Saleem RA, Rogers RS, Ratushny AV, Dilworth DJ, Shannon PT, Shteynberg D, Wan Y, Moritz RL, Nesvizhskii AI, Rachubinski RA, Aitchison JD. Saleem RA, et al. Mol Cell Proteomics. 2010 Sep;9(9):2076-88. doi: 10.1074/mcp.M000116-MCP201. Epub 2010 Apr 15. Mol Cell Proteomics. 2010. PMID: 20395639 Free PMC article. - Multiplex N-terminome analysis of MMP-2 and MMP-9 substrate degradomes by iTRAQ-TAILS quantitative proteomics.
Prudova A, auf dem Keller U, Butler GS, Overall CM. Prudova A, et al. Mol Cell Proteomics. 2010 May;9(5):894-911. doi: 10.1074/mcp.M000050-MCP201. Epub 2010 Mar 20. Mol Cell Proteomics. 2010. PMID: 20305284 Free PMC article.
References
- Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. - PubMed