Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype - PubMed (original) (raw)
Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype
K Mazan-Mamczarz et al. Oncogene. 2008.
Abstract
In a breast tumor xenograft model, the MCT-1 oncogene increases the in vivo tumorgenicity of MCF7 cells by promoting angiogenesis and inhibiting apoptosis. Increases in the tumor microvascular density are accompanied by a strong reduction in the levels of the angiogenesis inhibitor thrombospondin-1 (TSP1), but the mechanisms underlying this process are unknown. We show that TSP1 expression is controlled, at least in part, by post-transcriptional events. Using RNA interference to knock down the expression of the RNA-binding protein HuR in MCF7 cells as well as HuR overexpression, we demonstrate that HuR plays an important role in translation of the TSP1 mRNA. Furthermore, employing the RIP-Chip assay yielded 595 transcripts with significantly altered binding to HuR in the more tumorigenic breast cancer clones compared with the weakly tumorigenic clones. These mRNAs clustered in several pathways implicated in the transformed phenotype, such as the RAS pathway (involved in mitogenesis), the PI3K pathway (evasion of apoptosis) and pathways mediating angiogenesis and the cellular response to hypoxia. These findings demonstrate for the first time that global changes in HuR-bound mRNAs are implicated in the evolution to a more tumorigenic phenotype in an in vivo tumor model and underscore the role of global mRNA-protein interactions toward tumor progression.
Figures
Figure 1
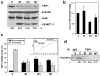
MCT-1 overexpression decreases translation and protein expression of TSP1 in MCF7 cells. (a) Representative Western blot analysis of cells stable transfected with the empty vector (V), and cells stable overexpressing MCT-1; mass culture (M) and to different clones (N1 and N7). 50 μg of total protein lysates were loaded and the abundance of TSP1, β-Actin, HuR and MCT-1 was assessed. (b) Changes in TSP1 total mRNA levels (relative to GAPDH mRNA) assessed by RT-qPCR for vector (V), and MCT-1 overexpressiong cells; mass culture (M) and two clones (N1 and N7). Graphs represent the means and standard errors of the means (SEM) from three independent experiments. Clone N1 showed less than 2-fold difference compared to vector (V); t-test value of p <0.01. (c) The relative association of the TSP1 mRNA with polysomes was tested by preparing cytoplasmic lysates from MCF7 stable cell lines (described in Fig. 1a), fractionating them through sucrose gradients and collecting 11 fraction for analysis. Representative polysome distribution profile obtained after centrifugation of cytopasmic lysates over sucrose gradient (inset); free mRNA, monosomes and polysomes of increasing molecular weight are indicated. The levels of TSP1 mRNA in the pooled 1–5 fractions comprised the nontranslating RNA, and pooled 6–11 fractions comprised the translating RNA were quantified by RT-qPCR and normalized to GAPDH mRNA. Graphs represent the means and SEM from six independent experiments; p <0.01 significantly different compared to vector (V) by t-test. (d) Newly translated TSP1 protein was assessed by 20 min incubation of each stable transfected MCF7 cell lines (described in Fig. 1a) with L-[35S]methionine and L-[35S]cysteine and immunoprecipitation (IP) of protein lysates with either anti-TSP1 antibody or control IgG. Samples were resolved by SDS-PAGE, transferred onto membranes and signal was visualized by PhosphoImager. Representative data are shown. In a and c, quantification of TSP1 signals is expressed as the percentage of the signal intensity relative to that in vector (V) cells.
Figure 2
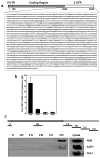
HuR binds the TSP1 mRNA. (a) Sequence of TSP1 mRNA depicting AREs sequences (underline) in TSP1 3′-untranslated region (3′UTR). (b) The association of TSP1 mRNA with ARE binding proteins (HuR, AUF1/HRNDP, TIA-1, and TIAR) was checked by immunoprecipitation (IP) of MCF7 cell lysates using either control (IgG) or specific antibodies. The obtained RNA was reverse transcribed and measured by qPCR. Shown are means and SEM from three independent experiments; GAPDH mRNA background amplification in the IP material was used as normalization control. (c) Schematic of TSP1 mRNA, representing the biotynolated transcripts (5′UTR, CR, and 3′UTR - A, B, C and D fragments) used for biotin pull-down analysis (top). Two ug of each biotinylated transcript were incubated with 40 μg of MCF7 cytoplasmic lysates. The presence of HuR, AUF1/HRNDP and TIA-1 in the pull-down material was detected by Western blotting; representative picture of couple repeats is shown.
Figure 3
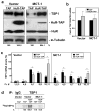
MCT-1 overexpression decreased the association of HuR with TSP1 mRNA. (a) Cytoplasmic lysates from stable transfected MCF7 cell lines (described in Fig. 1a) were employed for IP assays, using antibodies that recognized HuR, AUF1/hnRNP D, TIA-1 or TIAR. RNA was isolated and RT following qPCR was performed. Graph represents the mean and SEM from three independent assays. p < 0.02 versus vector (V) in IPs with HuR antibody. (b) Representative IP assays performed as described for Fig. 3a but followed by Western blotting analysis to assess the abundance of HuR protein in the IP material.
Figure 4
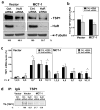
Overexpression of HuR in MCF7 cells increases translation and TSP1 protein expression. (a) MCF7 cells with normal (Vector) and overexpressing MCT-1 (MCT-1) levels were transiently transfected with either a control plasmid (TAP) or a plasmid overexpressing a tagged HuR (HuR-TAP), and lysate 72 hrs later. Ectopically expressed HuR (HuR-TAP), endogenous HuR, TSP1, and α-tubulin (loading control) levels were analyzed by Western blotting; representative data from three independent assays are shown. (b) The abundance of total TSP1 mRNA in cells expressing normal HuR levels (TAP) or overexpressing HuR (HuR-TAP) from both Vector and MCT-1 cell lines were measured by RT-qPCR in two separate experiments (mean values and SEM are shown). (c) MCF7 cells were transfected as in (a). mRNAs were extracted from each fraction, each of two or three consecutive fractions were pooled together (fractions 1, 2 and 3, fractions 4 and 5, fractions 6 and 7, fractions 8 and 9, fractions 10 and 11) and the TSP1 mRNAs were measured by RT-qPCR. Data were normalized to the GAPDH mRNA. Graphs represent the means and SEM from three independent experiments. *, p <0.05. (d) De novo TSP1 translation in MCF7 cell lines (Vector and overexpressing MCT-1) transfected as in (a) was assessed by cells incubation in L-[35S]methionine and L[35S]cysteine, IP reaction using anti-TSP1 antibody or IgG, protein electrophoresis by SDS-PAGE and detection of incorporated radiolabeled amino acids into newly synthesized TSP1 protein by PhosphorImager. Representative results are shown. In a and d, TSP1 signals were quantified and are represented as the percentage of the signal intensity in the empty vector (TAP) group.
Figure 5
HuR silencing reduces the expression of TSP1 in MCF7 cells. (a) MCF7 cells (Vector and overexpressing MCT-1) were transiently transfected either with control siRNA (Ctrl. siRNA) or siRNA targeting HuR (HuR siRNA). 72 hrs later Western blot analysis of HuR, TSP1, and the loading control α-tubulin was performed. Assays were repeated three times; representative pictures are shown. (b) RT-PCR analysis to monitor total TSP1 mRNA levels by 72 hrs after siRNA transfection. Analysis was performed three times independently. (c) 72 hrs after transfection cells with HuR-targeting (HuR siRNA) or control (Ctrl. siRNA) siRNAs, TSP1 mRNA levels in sucrose fractions were measured as described in legend of Fig. 4c. Graphs depict the means and SEM from three independent experiments. *, p < 0.05. (d) Changes in newly translated TSP1 in cells transfected as in (a) were measured as described for Fig. 4d. Shown are representative results from three separate assays.
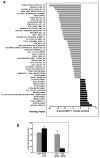
Figure 6
Influence of HuR on MCT-1-overexpressing MCF7 cells. (a) Top 48 biological pathways with the highest percentage of genes showing significantly altered association with HuR in response to overexpression of MCT-1 in MCF7 cells. Microarray analysis was performed in triplicate and the resulting data were normalized by Z score transformation and tested for significant differences in signal intensity. Differentially expressed genes (with Z ratio >1.5 or <-1.5, and fdr <0.3) were organized by the Ingenuity Pathway Analysis into known biological pathways. Probability scores for each functional grouping were calculated based on the chance of mRNA abundance changes predicting these interactions. The significance of each pathway was calculated by using the right-tailed Fisher’s Exact Test. The p-value was calculated by comparing the number of user-specified genes of interest involved in a given pathway relative to the total number of occurrences of these genes in all pathway annotations stored in the knowledge base. Graph shows pathways in which association with HuR significantly decreased with Z-score <−1.5 (gray bars) or increased with Z-score >1.5 (black bars), p <0.05. Pathways and genes involved are described in Table 1 and Supplementary information (Table S1). (b) Modulation of HuR levels alters anchorage-independent colony growth in MCT-1 overexpressing MCF7 cells. 24 h after transfection cells were cultured in agarose/medium, one week later colonies were counted. Graphs represent the mean and SEM of three independent experiments, p <0.03.
Similar articles
- Overexpression of the RNA binding protein HuR impairs tumor growth in triple negative breast cancer associated with deficient angiogenesis.
Gubin MM, Calaluce R, Davis JW, Magee JD, Strouse CS, Shaw DP, Ma L, Brown A, Hoffman T, Rold TL, Atasoy U. Gubin MM, et al. Cell Cycle. 2010 Aug 15;9(16):3337-46. doi: 10.4161/cc.9.16.12711. Epub 2010 Aug 17. Cell Cycle. 2010. PMID: 20724828 Free PMC article. - The RNA-binding protein HuR regulates GATA3 mRNA stability in human breast cancer cell lines.
Licata LA, Hostetter CL, Crismale J, Sheth A, Keen JC. Licata LA, et al. Breast Cancer Res Treat. 2010 Jul;122(1):55-63. doi: 10.1007/s10549-009-0517-8. Epub 2009 Sep 2. Breast Cancer Res Treat. 2010. PMID: 19728080 - The RNA binding protein HuR differentially regulates unique subsets of mRNAs in estrogen receptor negative and estrogen receptor positive breast cancer.
Calaluce R, Gubin MM, Davis JW, Magee JD, Chen J, Kuwano Y, Gorospe M, Atasoy U. Calaluce R, et al. BMC Cancer. 2010 Apr 6;10:126. doi: 10.1186/1471-2407-10-126. BMC Cancer. 2010. PMID: 20370918 Free PMC article. - HuR, a key post-transcriptional regulator, and its implication in progression of breast cancer.
Yuan Z, Sanders AJ, Ye L, Jiang WG. Yuan Z, et al. Histol Histopathol. 2010 Oct;25(10):1331-40. doi: 10.14670/HH-25.1331. Histol Histopathol. 2010. PMID: 20712017 Review. - Posttranscriptional orchestration of an anti-apoptotic program by HuR.
Abdelmohsen K, Lal A, Kim HH, Gorospe M. Abdelmohsen K, et al. Cell Cycle. 2007 Jun 1;6(11):1288-92. doi: 10.4161/cc.6.11.4299. Epub 2007 Jun 15. Cell Cycle. 2007. PMID: 17534146 Review.
Cited by
- RNA binding proteins (RBPs) and their role in DNA damage and radiation response in cancer.
Mehta M, Raguraman R, Ramesh R, Munshi A. Mehta M, et al. Adv Drug Deliv Rev. 2022 Dec;191:114569. doi: 10.1016/j.addr.2022.114569. Epub 2022 Oct 14. Adv Drug Deliv Rev. 2022. PMID: 36252617 Free PMC article. Review. - Hu Antigen R (HuR) Protein Structure, Function and Regulation in Hepatobiliary Tumors.
Lachiondo-Ortega S, Delgado TC, Baños-Jaime B, Velázquez-Cruz A, Díaz-Moreno I, Martínez-Chantar ML. Lachiondo-Ortega S, et al. Cancers (Basel). 2022 May 27;14(11):2666. doi: 10.3390/cancers14112666. Cancers (Basel). 2022. PMID: 35681645 Free PMC article. Review. - HuR function in disease.
Srikantan S, Gorospe M. Srikantan S, et al. Front Biosci (Landmark Ed). 2012 Jan 1;17(1):189-205. doi: 10.2741/3921. Front Biosci (Landmark Ed). 2012. PMID: 22201738 Free PMC article. Review. - Thrombospondin-1 is a critical effector of oncosuppressive activity of sst2 somatostatin receptor on pancreatic cancer.
Laklai H, Laval S, Dumartin L, Rochaix P, Hagedorn M, Bikfalvi A, Le Guellec S, Delisle MB, Schally AV, Susini C, Pyronnet S, Bousquet C. Laklai H, et al. Proc Natl Acad Sci U S A. 2009 Oct 20;106(42):17769-74. doi: 10.1073/pnas.0908674106. Epub 2009 Oct 1. Proc Natl Acad Sci U S A. 2009. PMID: 19805200 Free PMC article. - The complex world of post-transcriptional mechanisms: is their deregulation a common link for diseases? Focus on ELAV-like RNA-binding proteins.
Pascale A, Govoni S. Pascale A, et al. Cell Mol Life Sci. 2012 Feb;69(4):501-17. doi: 10.1007/s00018-011-0810-7. Epub 2011 Sep 10. Cell Mol Life Sci. 2012. PMID: 21909784 Free PMC article. Review.
References
- Aghib DF, Bishop JM, Ottolenghi S, Guerrasio A, Serra A, Saglio G. A 3′ truncation of MYC caused by chromosomal translocation in a human T-cell leukemia increases mRNA stability. Oncogene. 1990;5:707–711. - PubMed
- Bevilacqua A, Ceriani MC, Capaccioli S, Nicolin A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J Cell Physiol. 2003;195:356–372. - PubMed
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous