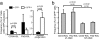Generation of mTert-GFP mice as a model to identify and study tissue progenitor cells - PubMed (original) (raw)
Generation of mTert-GFP mice as a model to identify and study tissue progenitor cells
David T Breault et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Stem cells hold great promise for regenerative medicine, but remain elusive in many tissues in part because universal markers of "stemness" have not been identified. The ribonucleoprotein complex telomerase catalyzes the extension of chromosome ends, and its expression is associated with failure of cells to undergo cellular senescence. Because such resistance to senescence is a common characteristic of many stem cells, we hypothesized that telomerase expression may provide a selective biomarker for stem cells in multiple tissues. In fact, telomerase expression has been demonstrated within hematopoietic stem cells. We therefore generated mouse telomerase reverse transcriptase (mTert)-GFP-transgenic mice and assayed the ability of mTert-driven GFP to mark tissue stem cells in testis, bone marrow (BM), and intestine. mTert-GFP mice were generated by using a two-step embryonic stem cell-based strategy, which enabled primary and secondary screening of stably transfected clones before blastocyst injection, greatly increasing the probability of obtaining mTert reporter mice with physiologically appropriate regulation of GFP expression. Analysis of adult mice showed that GFP is expressed in differentiating male germ cells, is enriched among BM-derived hematopoietic stem cells, and specifically marks long-term BrdU-retaining intestinal crypt cells. In addition, telomerase-expressing GFP(+) BM cells showed long-term, serial, multilineage BM reconstitution, fulfilling the functional definition of hematopoietic stem cells. Together, these data provide direct evidence that mTert-GFP expression marks progenitor cells in blood and small intestine, validating these mice as a useful tool for the prospective identification, isolation, and functional characterization of progenitor/stem cells from multiple tissues.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
_mTert_-GFP expression during EB formation. (a) Representative undifferentiated ES cell colony stably transfected with _mTert_-GFP. (b) Representative EB 7 days after LIF withdrawal. Note the outer layer of differentiated cells no longer expressing GFP. (c) Representative EB after 48 h of DMSO exposure and 7 days of LIF withdrawal. Note lack of GFP in the differentiated cell outgrowth. Each panel is a merge of Nomarski imaging (Upper Left Insets), DAPI stained nuclei (Upper Center Insets), and GFP expression (Upper Right Insets). (Magnification: ×20.)
Fig. 2.
mTert_-GFP expression in testis. (a) Western blotting was performed by using whole testis protein extract from WT, mTert_-GFP, or Actin-GFP mice and rabbit anti-GFP antibody. The blot was stripped and reblotted by using anti-Actin antibody as a loading control. (b) Representative FACS analysis for GFP expression performed on single-cell isolates from seminiferous tubules of adult mTert_-GFP and WT testes. Cells were gated into 1_N, 2_N, and 4_N populations. The dashed line illustrates the GFP threshold as defined by WT control cells. Histogram represents the mean ± SEM from two independent experiments with 3–5 replicates; ANOVA, P < 0.001, post hoc Fisher's (PLSD) analysis revealed significant differences among (*) groups (1_N_, 2_N_, and 4_N_) for both lines and between (#) the 2_N_ populations. (c) ISH for GFP expression in _mTert_-GFP adult testis. Dotted lines outline the seminiferous tubules. Arrowheads demarcate the basal layer of spermatogonial stem cells. (Left) Dark field images. (Right) Bright field images. (Magnification: ×10.) (d) Immunohistochemistry for GFP expression in _mTert_-GFP testis. Arrowhead indicates the basal layer of spermatogonial stem cells. (Magnification: Upper, ×10; Lower, ×40.)
Fig. 3.
Phenotypic Analysis of _mTert_-GFP expression in HSCs. (a) Increased frequency of LT HSCs and decreased frequency of myeloid progenitors among GFPhi populations by using multicolor FACS analysis of LT HSCs. Pooled results (mean ± SEM) from two independent experiments; Student's t test indicated. (b) Increased frequency of GFPhi cells within the LT HSC population compared with the ST HSC population. Pooled results from two independent experiments (mean ± SEM), Student's t test indicated.
Fig. 4.
Functional analysis of _mTert_-GFP expression in HSCs. (a) FACS scatter plot of BM from a male mTert_-GFP mouse demonstrating GFP+ cells above the dotted line (set using WT BM). GFP+ or GFP− cells were sorted according to the gates shown and transplanted into sublethally irradiated female recipient mice. (b) FACS analysis of BM from female recipient mice 5 months after transplant. GFP+ cells were sorted and transplanted into secondary female recipients. (c) FACS analysis of PBCs 5 months after serial BMT confirm serial LT engraftment of GFP+ cells. (d) TRAP assay was performed by using isolated GFP− and GFP+ BM cells (Fig. 4_a) to determine telomerase activity. Heat inactivation samples were used as a negative control. (e) FISH for Y chromosome demonstrated engraftment into both myeloid (Gr-1+ and Mac-1+) and lymphoid (B220+ and CD4+) lineages 2 months after serial BMT. (f) FACS analysis of GFP+ PBCs 5 months after transplantation confirm GFP in both myeloid (Mac-1+) and lymphoid (B220+, CD4+) lineages at levels comparable to donor animals.
Fig. 5.
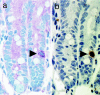
GFP expression in intestinal stem cells. Immunohistochemical analysis for GFP and BrdU using serial sections from _mTert_-GFP mice having undergone BrdU pulse–chase. (a) Staining with anti-GFP antibody, VIP (dark purple) chromagen substrate. (b) Staining with anti-BrdU antibody, DAB (dark brown) chromagen substrate. (Magnification: ×40.)
Similar articles
- The mouse endometrium contains epithelial, endothelial and leucocyte populations expressing the stem cell marker telomerase reverse transcriptase.
Deane JA, Ong YR, Cain JE, Jayasekara WS, Tiwari A, Carlone DL, Watkins DN, Breault DT, Gargett CE. Deane JA, et al. Mol Hum Reprod. 2016 Apr;22(4):272-84. doi: 10.1093/molehr/gav076. Epub 2016 Jan 5. Mol Hum Reprod. 2016. PMID: 26740067 Free PMC article. - Telomerase expression marks transitional growth-associated skeletal progenitor/stem cells.
Carlone DL, Riba-Wolman RD, Deary LT, Tovaglieri A, Jiang L, Ambruzs DM, Mead BE, Shah MS, Lengner CJ, Jaenisch R, Breault DT. Carlone DL, et al. Stem Cells. 2021 Mar;39(3):296-305. doi: 10.1002/stem.3318. Epub 2021 Jan 13. Stem Cells. 2021. PMID: 33438789 Free PMC article. - The proximal promoter region of mTert is sufficient to regulate telomerase activity in ES cells and transgenic animals.
Pericuesta E, Ramírez MA, Villa-Diaz A, Relaño-Gines A, Torres JM, Nieto M, Pintado B, Gutiérrez-Adán A. Pericuesta E, et al. Reprod Biol Endocrinol. 2006 Feb 3;4:5. doi: 10.1186/1477-7827-4-5. Reprod Biol Endocrinol. 2006. PMID: 16457732 Free PMC article. - Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells.
Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk ME, Henderson DE, Baffour-Awuah NY, Ambruzs DM, Fogli LK, Algra S, Breault DT. Montgomery RK, et al. Proc Natl Acad Sci U S A. 2011 Jan 4;108(1):179-84. doi: 10.1073/pnas.1013004108. Epub 2010 Dec 20. Proc Natl Acad Sci U S A. 2011. PMID: 21173232 Free PMC article. - Telomerase Reverse Transcriptase Expression in Mouse Endometrium During Reepithelialization and Regeneration in a Menses-Like Model.
Cousins FL, O DF, Ong YR, Breault DT, Deane JA, Gargett CE. Cousins FL, et al. Stem Cells Dev. 2019 Jan 1;28(1):1-12. doi: 10.1089/scd.2018.0133. Epub 2018 Nov 27. Stem Cells Dev. 2019. PMID: 30358490
Cited by
- Hallmarks of stemness in mammalian tissues.
Beumer J, Clevers H. Beumer J, et al. Cell Stem Cell. 2024 Jan 4;31(1):7-24. doi: 10.1016/j.stem.2023.12.006. Cell Stem Cell. 2024. PMID: 38181752 Free PMC article. Review. - Pancreatic stem/progenitor cells for the treatment of diabetes.
Noguchi H. Noguchi H. Rev Diabet Stud. 2010 Summer;7(2):105-11. doi: 10.1900/RDS.2010.7.105. Epub 2010 Aug 10. Rev Diabet Stud. 2010. PMID: 21060969 Free PMC article. Review. - Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice.
Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K. Arnold K, et al. Cell Stem Cell. 2011 Oct 4;9(4):317-29. doi: 10.1016/j.stem.2011.09.001. Cell Stem Cell. 2011. PMID: 21982232 Free PMC article. - The mouse endometrium contains epithelial, endothelial and leucocyte populations expressing the stem cell marker telomerase reverse transcriptase.
Deane JA, Ong YR, Cain JE, Jayasekara WS, Tiwari A, Carlone DL, Watkins DN, Breault DT, Gargett CE. Deane JA, et al. Mol Hum Reprod. 2016 Apr;22(4):272-84. doi: 10.1093/molehr/gav076. Epub 2016 Jan 5. Mol Hum Reprod. 2016. PMID: 26740067 Free PMC article. - Cell lineage identification and stem cell culture in a porcine model for the study of intestinal epithelial regeneration.
Gonzalez LM, Williamson I, Piedrahita JA, Blikslager AT, Magness ST. Gonzalez LM, et al. PLoS One. 2013 Jun 28;8(6):e66465. doi: 10.1371/journal.pone.0066465. Print 2013. PLoS One. 2013. PMID: 23840480 Free PMC article.
References
- Kondo M, et al. Biology of hematopoietic stem cells and progenitors: Implications for clinical application. Annu Rev Immunol. 2003;21:759–806. - PubMed
- Lovell-Badge R. The future for stem cell research. Nature. 2001;414:88–91. - PubMed
- Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. - PubMed
- Armstrong L, Lako M, Lincoln J, Cairns PM, Hole N. mTert expression correlates with telomerase activity during the differentiation of murine embryonic stem cells. Mech Dev. 2000;97:109–116. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5T32 DK007699/DK/NIDDK NIH HHS/United States
- R01 DK084056/DK/NIDDK NIH HHS/United States
- T32 DK007260/DK/NIDDK NIH HHS/United States
- R37 DK032658/DK/NIDDK NIH HHS/United States
- P30 HD018655/HD/NICHD NIH HHS/United States
- K08 DK066305/DK/NIDDK NIH HHS/United States
- P30 HD18655/HD/NICHD NIH HHS/United States
- R37 DK32658/DK/NIDDK NIH HHS/United States
- T32 DK007699/DK/NIDDK NIH HHS/United States
- 2T32 DK07260/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous