Mitogenomic evaluation of the historical biogeography of cichlids toward reliable dating of teleostean divergences - PubMed (original) (raw)
Mitogenomic evaluation of the historical biogeography of cichlids toward reliable dating of teleostean divergences
Yoichiro Azuma et al. BMC Evol Biol. 2008.
Abstract
Background: Recent advances in DNA sequencing and computation offer the opportunity for reliable estimates of divergence times between organisms based on molecular data. Bayesian estimations of divergence times that do not assume the molecular clock use time constraints at multiple nodes, usually based on the fossil records, as major boundary conditions. However, the fossil records of bony fishes may not adequately provide effective time constraints at multiple nodes. We explored an alternative source of time constraints in teleostean phylogeny by evaluating a biogeographic hypothesis concerning freshwater fishes from the family Cichlidae (Perciformes: Labroidei).
Results: We added new mitogenomic sequence data from six cichlid species and conducted phylogenetic analyses using a large mitogenomic data set. We found a reciprocal monophyly of African and Neotropical cichlids and their sister group relationship to some Malagasy taxa (Ptychochrominae sensu Sparks and Smith). All of these taxa clustered with a Malagasy + Indo/Sri Lankan clade (Etroplinae sensu Sparks and Smith). The results of the phylogenetic analyses and divergence time estimations between continental cichlid clades were much more congruent with Gondwanaland origin and Cretaceous vicariant divergences than with Cenozoic transmarine dispersal between major continents.
Conclusion: We propose to add the biogeographic assumption of cichlid divergences by continental fragmentation as effective time constraints in dating teleostean divergence times. We conducted divergence time estimations among teleosts by incorporating these additional time constraints and achieved a considerable reduction in credibility intervals in the estimated divergence times.
Figures
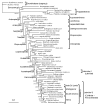
Figure 1
A Bayesian tree based on mitogenomic DNA sequences. This is a 50% majority rule consensus tree among 10,000 pooled trees from two independent Bayesian MCMC runs. The data set comprises aligned gap-free nucleotide sequences of 10,034-bp length from 54 taxa, which included 4,887 variable sites and 3,936 parsimony-informative sites. Partitioned Bayesian analyses were conducted using the GTR + I + Γ model and with all model parameters variable and unlinked across partitions. The numerals at internal nodes or branches indicate Bayesian posterior probabilities (left) and maximum likelihood bootstrap probability values (right) from 1000 replicates, respectively (shown as percentage for values above 50%).
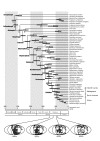
Figure 2
Divergence times estimated from the partitioned Bayesian analysis. A posterior distribution of divergence times with 95% credibility intervals (shaded rectangles) was obtained using mitogenomic DNA sequences (10,034 sites). Two sharks (Scyliorhinus canicula and Mustelus manazo) were used as an outgroup (not shown). The multidistribute program [41] was used to estimate divergence times assuming the tree topology shown in Fig. 1. Letters indicate nodes at which maximum and/or minimum time constraints were set (see Table 2 for details of the individual constraints). Paleogeographical maps at 148 MYA, 120 MYA, 95 MYA, and 85 MYA [50] are shown. Dark-gray areas on the maps represent those being fragmented within Gondwanaland at those times.
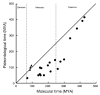
Figure 3
Comparison of paleontological and molecular estimates of divergence times. Minimum estimates of divergence times deducible from fossil records (see Table 2) were plotted as closed circles against molecularly estimated divergence times (mean values for the divergence times shown in Fig. 2). Closed triangles show plots of the timing of continental breakups against the molecular time estimates of cichlid divergences between the corresponding continents (data taken from Fig. 2). The timings used for complete continental breakups are 112 MYA for (Africa + South America) vs. (Madagascar + Indo/Sri Lanka), 100 MYA for Africa vs. South America, and 85 MYA for Madagascar vs. Indo/Sri Lanka [50-52]. The solid line indicates a 1:1 relationship between paleontological and molecular time estimates.
Figure 4
Divergence times estimated from the partitioned Bayesian analysis using both paleontological time constraints (Table 2) and biogeographical assumptions for the divergences of continental cichlid groups. The added time constraints on cichlid divergences are as follows: 112 MYA (lower) and 145 MYA (upper) for (Africa + South America) vs. (Madagascar + Indo/Sri Lanka); 100 MYA (lower) and 120 MYA (upper) for Africa vs. South America; and 85 MYA (lower) and 95 MYA (upper) for Madagascar vs. Indo/Sri Lanka [50-52]. See Fig. 2 legend for other details.
Similar articles
- Molecular phylogeny and biogeography of the Malagasy and South Asian cichlids (Teleostei: Perciformes: Cichlidae).
Sparks JS. Sparks JS. Mol Phylogenet Evol. 2004 Mar;30(3):599-614. doi: 10.1016/S1055-7903(03)00225-2. Mol Phylogenet Evol. 2004. PMID: 15012941 - Bayesian Phylogenetic Estimation of Clade Ages Supports Trans-Atlantic Dispersal of Cichlid Fishes.
Matschiner M, Musilová Z, Barth JM, Starostová Z, Salzburger W, Steel M, Bouckaert R. Matschiner M, et al. Syst Biol. 2017 Jan 1;66(1):3-22. doi: 10.1093/sysbio/syw076. Syst Biol. 2017. PMID: 28173588 - Multilocus phylogeny and rapid radiations in Neotropical cichlid fishes (Perciformes: Cichlidae: Cichlinae).
López-Fernández H, Winemiller KO, Honeycutt RL. López-Fernández H, et al. Mol Phylogenet Evol. 2010 Jun;55(3):1070-86. doi: 10.1016/j.ympev.2010.02.020. Epub 2010 Feb 21. Mol Phylogenet Evol. 2010. PMID: 20178851 - Molecular and fossil evidence place the origin of cichlid fishes long after Gondwanan rifting.
Friedman M, Keck BP, Dornburg A, Eytan RI, Martin CH, Hulsey CD, Wainwright PC, Near TJ. Friedman M, et al. Proc Biol Sci. 2013 Sep 18;280(1770):20131733. doi: 10.1098/rspb.2013.1733. Print 2013 Nov 7. Proc Biol Sci. 2013. PMID: 24048155 Free PMC article. - Genetic Variation and Hybridization in Evolutionary Radiations of Cichlid Fishes.
Svardal H, Salzburger W, Malinsky M. Svardal H, et al. Annu Rev Anim Biosci. 2021 Feb 16;9:55-79. doi: 10.1146/annurev-animal-061220-023129. Epub 2020 Nov 16. Annu Rev Anim Biosci. 2021. PMID: 33197206 Review.
Cited by
- Trophic divergence of Lake Kivu cichlid fishes along a pelagic versus littoral habitat axis.
Munyandamutsa PS, Jere WL, Kassam D, Mtethiwa A. Munyandamutsa PS, et al. Ecol Evol. 2021 Jan 27;11(4):1570-1585. doi: 10.1002/ece3.7117. eCollection 2021 Feb. Ecol Evol. 2021. PMID: 33613990 Free PMC article. - The genomic timeline of cichlid fish diversification across continents.
Matschiner M, Böhne A, Ronco F, Salzburger W. Matschiner M, et al. Nat Commun. 2020 Nov 18;11(1):5895. doi: 10.1038/s41467-020-17827-9. Nat Commun. 2020. PMID: 33208747 Free PMC article. - Rare coral under the genomic microscope: timing and relationships among Hawaiian Montipora.
Cunha RL, Forsman ZH, Belderok R, Knapp ISS, Castilho R, Toonen RJ. Cunha RL, et al. BMC Evol Biol. 2019 Jul 24;19(1):153. doi: 10.1186/s12862-019-1476-2. BMC Evol Biol. 2019. PMID: 31340762 Free PMC article. - New fossil cichlid from the middle Miocene of East Africa revealed as oldest known member of the Oreochromini.
Penk SBR, Altner M, F Cerwenka A, Schliewen UK, Reichenbacher B. Penk SBR, et al. Sci Rep. 2019 Jul 15;9(1):10198. doi: 10.1038/s41598-019-46392-5. Sci Rep. 2019. PMID: 31308387 Free PMC article. - Osteology and phylogeny of Robustichthys luopingensis, the largest holostean fish in the Middle Triassic.
Xu GH. Xu GH. PeerJ. 2019 Jun 24;7:e7184. doi: 10.7717/peerj.7184. eCollection 2019. PeerJ. 2019. PMID: 31275762 Free PMC article.
References
- Yang Z. Computational Molecular Evolution. New York: Oxford University Press; 2006.
- Benton MJ. The Fossil Record. Vol. 2. London: Chapman & Hall; 1993.
- Kumazawa Y, Yamaguchi M, Nishida M. In: The biology of biodiversity. Kato M, editor. Tokyo: Springer; 1999. Mitochondrial molecular clocks and the origin of euteleostean biodiversity: Familial radiation of perciforms may have predated the Cretaceous/Tertiary boundary; pp. 35–52.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials