Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A - PubMed (original) (raw)
Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A
Alexander W Bird et al. J Cell Biol. 2008.
Abstract
To assemble mitotic spindles, cells nucleate microtubules from a variety of sources including chromosomes and centrosomes. We know little about how the regulation of microtubule nucleation contributes to spindle bipolarity and spindle size. The Aurora A kinase activator TPX2 is required for microtubule nucleation from chromosomes as well as for spindle bipolarity. We use bacterial artificial chromosome-based recombineering to introduce point mutants that block the interaction between TPX2 and Aurora A into human cells. TPX2 mutants have very short spindles but, surprisingly, are still bipolar and segregate chromosomes. Examination of microtubule nucleation during spindle assembly shows that microtubules fail to nucleate from chromosomes. Thus, chromosome nucleation is not essential for bipolarity during human cell mitosis when centrosomes are present. Rather, chromosome nucleation is involved in spindle pole separation and setting spindle length. A second Aurora A-independent function of TPX2 is required to bipolarize spindles.
Figures
Figure 1.
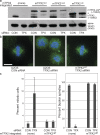
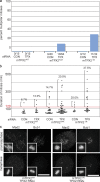
A BAC transgene containing mTPX2-LAP rescues the phenotype of TPX2-RNAi. (a) Western blot of cell lines with anti-mTPX2 antibody, which recognizes both human TPX2 and mTPX2-LAP, and anti–α-tubulin, after TPX2 or CON RNAi. hTPX2 is efficiently depleted in all lines after TPX2 RNAi, whereas mTPX2-LAP transgenes remain. The asterisk represents the position of the TPX2ΔN protein, which runs faster than the full length, as expected. (b) Immunofluorescence of U2OS cells after CON or TPX2 RNAi and TPX2WT cells after TPX2 RNAi, stained for α-tubulin (green), Cep135 (red), and DNA (blue). U2OS cells without a rescuing transgene show a characteristic phenotype after hTPX2 depletion of collapsed poles and lack of a bipolar spindle, whereas TPX2WT cells containing mTPX2-LAP after hTPX2 depletion have normal spindle morphology. Bar,10 μm. (c) Quantification of the TPX2 depletion phenotype and mTPX2-LAP rescue displayed in b. The percent of mitotic cells (n = >1,000 cells per experiment; three to five experiments for each condition) and percent bipolar spindles (n > 50) mitotic cells per experiment; three to five experiments per condition) after CON or TPX2 RNAi in U2OS or TPX2WT cells were determined. Error bars represent standard deviation. The mTPX2-LAP construct is able to rescue depletion of endogenous TPX2.
Figure 2.
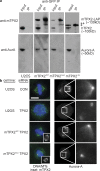
TPX2 mutants abolish the in vivo interaction between TPX2 and Aurora A as well as Aurora A localization to spindles. (a) Western blots of cell extracts immunoprecipitated to detect interaction of TPX2 with Aurora A. U2OS (untagged), mTPX2WT, mTPX2AAA, and mTPX2ΔN cells were arrested in mitosis with nocodazole, and protein extracts were immunoprecipitated with anti-GFP antibody. Input and immunoprecipitated fractions were run by SDS-PAGE and blotted with either anti-mTPX2 or anti–Aurora A antibody. Anti-GFP antibody pulls down mTPX2WT bound to Aurora A, whereas nothing is immunoprecipitated in the untagged cells. Immunoprecipitating mTPX2WT does not pull down endogenous TPX2. In mTPX2AAA and mTPX2ΔN cells, the mTPX2 mutant transgene is pulled down, but Aurora A is not. The single asterisk represents the position of the TPX2ΔN protein. The double asterisk represents a degradation product of the mTPX2 protein. (b) Immunofluorescence analysis of cell lines after CON or TPX2 RNAi stained for α-tubulin (green), DNA (blue), mTPX2-LAP (anti-GFP; insets), and Aurora A. Aurora A is localized to spindles and centrosomes in U2OS cells (top row). After hTPX2 RNAi, Aurora A is absent from spindles but still on centrosomes (second row). A mouse TPX2-LAP transgene restores Aurora A localization to spindles after hTPX2 RNAi (third row). The same mouse transgene with point mutations introduced to abolish the TPX2–Aurora A interaction no longer is able to recruit Aurora A to spindles after hTPX2 RNAi (bottom row). The pericentrosomal Aurora A localization shown in the mutant similar to that of the TPX2 depletion was observed in images of 12 of 14 cells. In 2 of 14 images, the Aurora A localization in the mutant reflected a more centrosomal localization. Bar, 10 μm.
Figure 3.
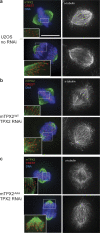
TPX2 localizes to kinetochore fibers independent of interaction with Aurora A. Immunofluorescence analysis of TPX2 localization. (a) U2OS cells in metaphase (top row) and anaphase (bottom row) stained for DNA, CREST, hTPX2 (green), and α-tubulin (black and white). Insets are enlarged to show TPX2 localization to microtubules that end at kinetochores (kinetochore fibers). (b) mTPX2WT cells after hTPX2(RNAi) with same staining as in a, except with mTPX2 instead of hTPX2 to recognize the transgene. (c) mTPX2AAA cells after hTPX2(RNAi) with same staining as in b. mTPX2 mutated to abolish interaction with Aurora A is still able to localize to kinetochore-fibers. Bar, 10 μm.
Figure 4.
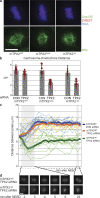
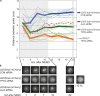
The TPX2–Aurora A interaction is required for spindle pole separation and spindle length establishment. (a) Immunofluorescence of mTPX2WT, mTPX2AAA, and mTPX2ΔN mitotic cells after hTPX2(RNAi), stained for CREST, Cep135, DNA, and α-tubulin. Centrosomes are collapsed to chromatin when the TPX2–Aurora A interaction is abolished. Bar, 10 μm. (b) Centrosome-kinetochore distances are shorter in mTPX2AAA and mTPX2ΔN mutants. The centrosome-kinetochore distance from cells stained as in a was determined by measuring individual centrosome-kinetochore distances from metaphase cells in three dimensions (see Materials and methods). For each condition, measurements were taken from four independent cells (eight centrosomes). Red dots show the data points measured and the bars show the means. (c) In TPX2 mutants unable to bind or activate Aurora A, spindle poles collapse immediately after NEBD and only slightly elongate to form shorter metaphase spindles. Distances between spindle poles were measured in three dimensions from live-cell image stacks taken at 1-min intervals of mTPX2WT, mTPX2AAA, and mTPX2ΔN (mTPX2-GFP) cells after hTPX2(RNAi). Thick lines represent means and thin lines represent individual videos. (d) Representative still images from time-lapse recordings quantified in b.
Figure 5.
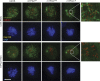
TPX2 mutants have a higher percentage of defective divisions, delayed mitoses, and increased metaphase BUB1 staining. (a) Percentage of defective mitoses in mTPX2WT, mTPX2AAA, and mTPX2ΔN cells after TPX2 or CON RNAi, quantified by observing time-lapse videos of GFP fluorescence. Actual numbers of cell divisions observed are shown below the histogram. (b) mTPX2AAA and mTPX2ΔN cells after TPX2 RNAi show longer delays in mitosis and a higher percentage of cells exhibiting delays. Lengths of individual mitosis are plotted for mTPX2WT, mTPX2AAA, and mTPX2ΔN cells after TPX2 or CON RNAi. Percentages shown are the fraction of cells showing lengths of mitoses greater than 120 min. (c) Immunofluorescence of mTPX2WT and mTPX2ΔN prometaphase and metaphase cells after TPX2 RNAi, stained for BUB1 or MAD2. Insets show DNA (DAPI) from the same cell at lower magnification. In mTPX2WT cells, MAD2 and BUB1 foci are enriched in prophase but not metaphase. In mTPX2ΔN cells, MAD2 enrichment is gone in metaphase cells, whereas some Bub1-positive foci remain. Bars, 10 μm.
Figure 6.
Depletion of TPX2 from cells results in a spindle defect phenotype distinct from TPX2 mutants or Aurora A depletion. (a) Distances between spindle poles were measured in three dimensions from live-cell image stacks taken at 3-min intervals of U2OS cells stably transfected with α-tubulin–mCherry after treatment with CON, TPX2, or AurA siRNA. Thick lines represent means and thin lines represent individual videos. The dashed line represents data from Fig. 4 for reference. (b) Representative still images from time-lapse recordings quantified in a. The cells shown transfected with CON or AurA siRNAs completed mitosis and divided, whereas the cell depleted of TPX2 arrested in mitosis for several hours and displayed fragmented spindle poles. Bar, 10 μm.
Figure 7.
The TPX2–Aurora A interaction is required for cold-stable microtubule stability and organization in prometaphase. mTPX2WT, mTPX2AAA, and mTPX2ΔN cells were transfected with TPX2 or CON siRNA. 48 h after transfection, cells were treated on ice for 5 min and fixed. Fluorescence images are shown of cells stained with anti–α-tubulin (microtubules) and DAPI (DNA) showing cold-stable microtubule populations. Bar, 10 μm.
Figure 8.
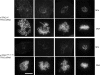
The TPX2–Aurora A interaction is required for chromatin/kinetochore-mediated microtubule nucleation. U2OS, mTPX2WT, mTPX2AAA, and mTPX2ΔN cells transfected with TPX2 or CON siRNA were treated on ice to completely depolymerize microtubules, transferred to 37°C for 90 s to allow repolymerization, and immediately fixed. Immunofluorescence images are shown of cells stained with α-tubulin and CREST (top rows) and Cep135 and DNA (bottom rows). In addition to aster microtubule polymerization associated with centrosomes, microtubule polymerization broadly associated with chromatin and often specifically associated with kinetochores was evident in all cell lines after CON siRNA and in mTPX2WT cells after TPX2 RNAi. In U2OS, mTPX2AAA, and mTPX2ΔN cells after TPX2 RNAi, microtubule repolymerization was no longer seen associated with chromatin or kinetochores, but centrosome-associated microtubule aster polymerization was still evident. Bar, 10 μm.
Similar articles
- A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association.
De Luca M, Lavia P, Guarguaglini G. De Luca M, et al. Cell Cycle. 2006 Feb;5(3):296-303. doi: 10.4161/cc.5.3.2392. Epub 2006 Feb 7. Cell Cycle. 2006. PMID: 16418575 - Characterization of the TPX2 domains involved in microtubule nucleation and spindle assembly in Xenopus egg extracts.
Brunet S, Sardon T, Zimmerman T, Wittmann T, Pepperkok R, Karsenti E, Vernos I. Brunet S, et al. Mol Biol Cell. 2004 Dec;15(12):5318-28. doi: 10.1091/mbc.e04-05-0385. Epub 2004 Sep 22. Mol Biol Cell. 2004. PMID: 15385625 Free PMC article. - The nuclear scaffold protein SAF-A is required for kinetochore-microtubule attachment and contributes to the targeting of Aurora-A to mitotic spindles.
Ma N, Matsunaga S, Morimoto A, Sakashita G, Urano T, Uchiyama S, Fukui K. Ma N, et al. J Cell Sci. 2011 Feb 1;124(Pt 3):394-404. doi: 10.1242/jcs.063347. J Cell Sci. 2011. PMID: 21242313 - Regulation of Aurora-A kinase on the mitotic spindle.
Kufer TA, Nigg EA, Silljé HH. Kufer TA, et al. Chromosoma. 2003 Dec;112(4):159-63. doi: 10.1007/s00412-003-0265-1. Epub 2003 Nov 21. Chromosoma. 2003. PMID: 14634755 Review. - TPX2: of spindle assembly, DNA damage response, and cancer.
Neumayer G, Belzil C, Gruss OJ, Nguyen MD. Neumayer G, et al. Cell Mol Life Sci. 2014 Aug;71(16):3027-47. doi: 10.1007/s00018-014-1582-7. Epub 2014 Feb 21. Cell Mol Life Sci. 2014. PMID: 24556998 Free PMC article. Review.
Cited by
- Interaction proteomics identify NEURL4 and the HECT E3 ligase HERC2 as novel modulators of centrosome architecture.
Al-Hakim AK, Bashkurov M, Gingras AC, Durocher D, Pelletier L. Al-Hakim AK, et al. Mol Cell Proteomics. 2012 Jun;11(6):M111.014233. doi: 10.1074/mcp.M111.014233. Epub 2012 Jan 19. Mol Cell Proteomics. 2012. PMID: 22261722 Free PMC article. - Intracellular Scaling Mechanisms.
Reber S, Goehring NW. Reber S, et al. Cold Spring Harb Perspect Biol. 2015 Aug 7;7(12):a019067. doi: 10.1101/cshperspect.a019067. Cold Spring Harb Perspect Biol. 2015. PMID: 26254310 Free PMC article. Review. - Centrobin controls primary ciliogenesis in vertebrates.
Ogungbenro YA, Tena TC, Gaboriau D, Lalor P, Dockery P, Philipp M, Morrison CG. Ogungbenro YA, et al. J Cell Biol. 2018 Apr 2;217(4):1205-1215. doi: 10.1083/jcb.201706095. Epub 2018 Feb 13. J Cell Biol. 2018. PMID: 29440264 Free PMC article. - Kinesin-12 Kif15 targets kinetochore fibers through an intrinsic two-step mechanism.
Sturgill EG, Das DK, Takizawa Y, Shin Y, Collier SE, Ohi MD, Hwang W, Lang MJ, Ohi R. Sturgill EG, et al. Curr Biol. 2014 Oct 6;24(19):2307-13. doi: 10.1016/j.cub.2014.08.022. Epub 2014 Sep 25. Curr Biol. 2014. PMID: 25264249 Free PMC article. - Aurora A phosphorylation of WD40-repeat protein 62 in mitotic spindle regulation.
Lim NR, Yeap YY, Ang CS, Williamson NA, Bogoyevitch MA, Quinn LM, Ng DC. Lim NR, et al. Cell Cycle. 2016;15(3):413-24. doi: 10.1080/15384101.2015.1127472. Cell Cycle. 2016. PMID: 26713495 Free PMC article.
References
- Barr, A.R., and F. Gergely. 2007. Aurora-A: the maker and breaker of spindle poles. J. Cell Sci. 120:2987–2996. - PubMed
- Basto, R., J. Lau, T. Vinogradova, A. Gardiol, C.G. Woods, A. Khodjakov, and J.W. Raff. 2006. Flies without centrioles. Cell. 125:1375–1386. - PubMed
- Bayliss, R., T. Sardon, I. Vernos, and E. Conti. 2003. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell. 12:851–862. - PubMed
- Bischoff, F.R., and H. Ponstingl. 1991. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 354:80–82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous