Comparative analysis of human gut microbiota by barcoded pyrosequencing - PubMed (original) (raw)
Comparative Study
Comparative analysis of human gut microbiota by barcoded pyrosequencing
Anders F Andersson et al. PLoS One. 2008.
Abstract
Humans host complex microbial communities believed to contribute to health maintenance and, when in imbalance, to the development of diseases. Determining the microbial composition in patients and healthy controls may thus provide novel therapeutic targets. For this purpose, high-throughput, cost-effective methods for microbiota characterization are needed. We have employed 454-pyrosequencing of a hyper-variable region of the 16S rRNA gene in combination with sample-specific barcode sequences which enables parallel in-depth analysis of hundreds of samples with limited sample processing. In silico modeling demonstrated that the method correctly describes microbial communities down to phylotypes below the genus level. Here we applied the technique to analyze microbial communities in throat, stomach and fecal samples. Our results demonstrate the applicability of barcoded pyrosequencing as a high-throughput method for comparative microbial ecology.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
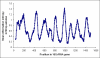
Figure 1. Variability within the 16S rRNA gene.
From pre-aligned sequenced >1200 bp downloaded from RDP, the variability, measured as Shannon information entropy, was calculated at each sequence position, using only positions without a gap in E. coli. The graph shows the Shannon entropy (y-axis) averaged over 50 bp windows, centered at each position in the gene (x-axis). Shannon entropy at position x was calculated as –Σ p(xi) log_2 p_(xi), where p(xi) denotes the frequency of nucleotide i. The filled arrows indicate positions of the PCR primers, the dashed arrow the direction of sequencing.
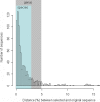
Figure 2. Taxonomic classification accuracy.
Distribution of sequence distances (measured over the whole sequence lengths) between original sequence and the selected reference sequence, when 59 bp corresponding to minimal pyrosequencing reads were extracted from 1000 randomly selected RDP sequences and assigned to reference RDP sequences according to the procedure described in the Materials and Methods section (in this case the 1000 sequences had first been removed from the BLAST database).
Figure 3. Comparison of the throat, stomach and fecal microbiotas.
a, A neighbor joining phylogenetic tree of the RDP sequences representing the 454 reads from six samples of throat, stomach, and feces, respectively, was constructed. Branches in the tree represented in throat, stomach, and feces are labeled with green, yellow, and red, respectively. b, Hierchical clustering of the 18 samples based on how their reads were distributed within the tree using the weighted UniFrac metric for pair wise comparisons of the samples. The lower three samples are H. pylori positive stomachs.
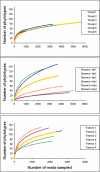
Figure 4. Rarefaction analysis of the different gut ecosystems.
Number of phylotypes sampled as a function of number of reads. The data points represent averages of 1000 randomized samplings without replacements.
References
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. - PubMed
- Wostmann BS, Larkin C, Moriarty A, Bruckner-Kardoss E. Dietary intake, energy metabolism, and excretory losses of adult male germfree Wistar rats. Lab Anim Sci. 1983;33:46–50. - PubMed
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. - PubMed
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases