Whole chromosome instability and cancer: a complex relationship - PubMed (original) (raw)
Review
Whole chromosome instability and cancer: a complex relationship
Robin M Ricke et al. Trends Genet. 2008 Sep.
Abstract
Although chromosome mis-segregation is a hallmark of cancer cells, its genetic basis and role in malignant transformation remain poorly understood. In recent years, several mouse models have been generated that harbor gene defects that perturb high-fidelity chromosome segregation. Analysis of these models has revealed that whole chromosome instability (W-CIN) can cause, inhibit or have no effect on tumorigenesis. Here we propose that the effect of W-CIN on tumor development depends on the particular W-CIN gene that is defective, including its other cellular functions, the extent or nature of the gene defect, the affected tissue or cell type and the context of other cancer gene mutations.
Figures
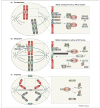
Figure 1
Current model of the mitotic checkpoint. To prevent chromosome mis-segregation, cells have developed a multiprotein surveillance mechanism, called the mitotic checkpoint, that delays anaphase onset until all kinetochores are correctly attached to mitotic spindle microtubules and aligned in the metaphase plate (the central area between the two spindle poles). (a) Various mitotic checkpoint proteins, including Rae1 (green), Bub3 (pink), Bub1 (orange), BubR1 (purple), Mad1 (blue) and Mad2 (yellow), bind kinetochores (green) that lack attachment or tension and generate a ‘stop anaphase’ signal that diffuses into the mitotic cytosol. This signal is believed to consist of Bub3–BubR1–Mad2 and Rae1–Nup98 (light yellow) protein complexes, which bind and inhibit APCCdc20 (dark grey) and APCCdh1 (light grey), respectively. Once each pair of sister kinetochores attaches to microtubules (black), and microtubule motors generate tension that stretches them, the production of inhibitory ‘stop anaphase’ signals at those kinetochores quenches. (b) Silencing of the ‘stop anaphase’ signal at the last kinetochore triggers the release of Bub3–BubR1–Mad2 and Rae1–Nup98 from APCCdc20 and APCCdh1, respectively. This event allows APCCdc20-mediated cyclin B (blue) destruction and APCCdh1-mediated securin (brown) degradation. Separase (grey), which is inhibited through its association with securin and by cyclin B–Cdk1-mediated phosphorylation, triggers sister chromatid disjunction by cohesin cleavage. Sister chromatid separation is also dependent on Topo IIα (grey), which decatenates centromeric DNA. Topo IIα targeting to inner centromeres is dependent on RanBP2 (purple)-mediated sumoylation. (c) In anaphase, fully separated sister chromatids move to opposite poles. Proper progression through anaphase requires APC/CCdh1-mediated degradation of Cdc20 and Polo like kinase 1 (Plk1; green). AurkA (yellow) and AurkB (pink) remain intact until their degradation in late mitosis. Blue ovals in (c) represent portions of the contractile ring that mediates cell division. Encircled C, cohesin; encircled U, ubiquitin.
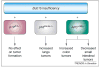
Figure 2
Genetic context of whole chromosome instability (W-CIN) genes modulates tumorigenesis propensity. The genetic context of W-CIN defects dramatically alters the tumorigenetic potential outcome of a given tissue type. As a specific example, Bub1b hypomorphism has been analyzed in mice lacking either p16Ink4a or p19Arf 47. Tumorigenesis was accelerated only in the context of p16Ink4a, but not p19Arf, loss. Similarly, APC+/Min mice develop more colon tumors, and fewer small intestine tumors, on a Bub1b+/− background than on a Bub1b+/+ background.
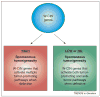
Figure 3
Differences among whole chromosome instability (W-CIN) genes in their tumor suppressive activities. Defects in some W-CIN genes are more probable than others to promote transformation. Based on the currently available data, we hypothesize that two broad classes of W-CIN genes exist. Defects in W-CIN genes that result in high spontaneous tumorigenecity (red box) might activate multiple tumor-promoting activities. Examples of such defects are: Bub1 hypomorphism (causes aneuploidy and promotes cell survival); RanBP2 hypomorphism (causes aneuploidy and perhaps aberrant nucleocytoplasmic transport) and APC inactivation (causes aneuploidy and uncontrolled cell proliferation through β-catenin signaling). We also suggest that defects in W-CIN genes that have either little or no effect on spontaneous tumorigenecity (green box) might, when deregulated, activate both tumor-promoting and anti-tumor activities. For example, Bub1b hypomorphism causes aneuploidy but also induces senescence, a cellular mechanism that protects against tumorigenesis.
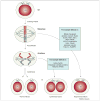
Figure 4
Tetraploidization by mitotic failure. Normally a 4N cell in G2 enters mitosis, aligns its chromosomes in the metaphase plate and equally distributes the DNA over two nuclei (karyokinesis) and subsequently two daughter cells (cytokinesis). When karyokinesis fails, cells with 4N content are observed. When cytokinesis fails, the DNA is divided into two nuclei that remain within one cell. Mitotic genes that could have a role in failure of karyokinesis and cytokinesis are indicated (blue boxes). Note that cells undergoing karyokinesis and cytokinesis both inherit two centrosomes that could lead to abnormal spindles and chromosome mis-segregation during the next round of division (Box 1).
Similar articles
- Chromosomal instability suppresses the growth of K-Ras-induced lung adenomas.
Laucius CD, Orr B, Compton DA. Laucius CD, et al. Cell Cycle. 2019 Aug;18(15):1702-1713. doi: 10.1080/15384101.2019.1629790. Epub 2019 Jun 24. Cell Cycle. 2019. PMID: 31179849 Free PMC article. - Chromosomal instability: A common feature and a therapeutic target of cancer.
Tanaka K, Hirota T. Tanaka K, et al. Biochim Biophys Acta. 2016 Aug;1866(1):64-75. doi: 10.1016/j.bbcan.2016.06.002. Epub 2016 Jun 21. Biochim Biophys Acta. 2016. PMID: 27345585 Review. - Unraveling pathologies underlying chromosomal instability in cancers.
Jo M, Kusano Y, Hirota T. Jo M, et al. Cancer Sci. 2021 Aug;112(8):2975-2983. doi: 10.1111/cas.14989. Epub 2021 Jun 11. Cancer Sci. 2021. PMID: 34032342 Free PMC article. Review. - The cancer biology of whole-chromosome instability.
Duijf PH, Benezra R. Duijf PH, et al. Oncogene. 2013 Oct;32(40):4727-36. doi: 10.1038/onc.2012.616. Epub 2013 Jan 14. Oncogene. 2013. PMID: 23318433 Review. - Increased replication origin firing links replication stress to whole chromosomal instability in human cancer.
Böhly N, Schmidt AK, Zhang X, Slusarenko BO, Hennecke M, Kschischo M, Bastians H. Böhly N, et al. Cell Rep. 2022 Dec 13;41(11):111836. doi: 10.1016/j.celrep.2022.111836. Cell Rep. 2022. PMID: 36516748
Cited by
- Let's huddle to prevent a muddle: centrosome declustering as an attractive anticancer strategy.
Ogden A, Rida PC, Aneja R. Ogden A, et al. Cell Death Differ. 2012 Aug;19(8):1255-67. doi: 10.1038/cdd.2012.61. Epub 2012 Jun 1. Cell Death Differ. 2012. PMID: 22653338 Free PMC article. Review. - Cdc20 is critical for meiosis I and fertility of female mice.
Jin F, Hamada M, Malureanu L, Jeganathan KB, Zhou W, Morbeck DE, van Deursen JM. Jin F, et al. PLoS Genet. 2010 Sep 30;6(9):e1001147. doi: 10.1371/journal.pgen.1001147. PLoS Genet. 2010. PMID: 20941357 Free PMC article. - Mad1 destabilizes p53 by preventing PML from sequestering MDM2.
Wan J, Block S, Scribano CM, Thiry R, Esbona K, Audhya A, Weaver BA. Wan J, et al. Nat Commun. 2019 Apr 4;10(1):1540. doi: 10.1038/s41467-019-09471-9. Nat Commun. 2019. PMID: 30948704 Free PMC article. - CAMP (C13orf8, ZNF828) is a novel regulator of kinetochore-microtubule attachment.
Itoh G, Kanno S, Uchida KS, Chiba S, Sugino S, Watanabe K, Mizuno K, Yasui A, Hirota T, Tanaka K. Itoh G, et al. EMBO J. 2011 Jan 5;30(1):130-44. doi: 10.1038/emboj.2010.276. Epub 2010 Nov 9. EMBO J. 2011. PMID: 21063390 Free PMC article. - BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase.
Malureanu LA, Jeganathan KB, Hamada M, Wasilewski L, Davenport J, van Deursen JM. Malureanu LA, et al. Dev Cell. 2009 Jan;16(1):118-31. doi: 10.1016/j.devcel.2008.11.004. Dev Cell. 2009. PMID: 19154723 Free PMC article.
References
- Boveri T. Zur Frage der Entstehung Maligner Tumoren. Gustav Fisher; 1914.
- Boveri T. Uber mehrpolige Mitosen als Mittel zur Analyse der Zelkerns. Vehr d phys Med Ges zu Wurzburg Neu Folge. 1902;35:67–90.
- Duesberg P, Li R. Multistep carcinogenesis: a chain reaction of aneuploidizations. Cell Cycle. 2003;2:202–210. - PubMed
- Zimonjic D, et al. Derivation of human tumor cells in vitro without widespread genomic instability. Cancer Res. 2001;61:8838–8844. - PubMed
- Michor F, et al. Can chromosomal instability initiate tumorigenesis? Semin Cancer Biol. 2005;15:43–49. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 CA096985/CA/NCI NIH HHS/United States
- R01 CA126828-02/CA/NCI NIH HHS/United States
- CA-126828/CA/NCI NIH HHS/United States
- CA-77262/CA/NCI NIH HHS/United States
- R01 CA077262/CA/NCI NIH HHS/United States
- R01 CA096985-07/CA/NCI NIH HHS/United States
- R01 CA126828/CA/NCI NIH HHS/United States
- CA-96985/CA/NCI NIH HHS/United States
- R01 CA077262-11/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases