AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development - PubMed (original) (raw)
. 2008 Sep 1;205(9):1949-57.
doi: 10.1084/jem.20081007. Epub 2008 Aug 4.
Helena Tolarová, Zhiyu Li, Wendy Dubois, Susan Lim, Elsa Callen, Sonia Franco, Maria Mosaico, Lionel Feigenbaum, Frederick W Alt, André Nussenzweig, Michael Potter, Rafael Casellas
Affiliations
- PMID: 18678733
- PMCID: PMC2526190
- DOI: 10.1084/jem.20081007
AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development
Makiko Takizawa et al. J Exp Med. 2008.
Abstract
Immunoglobulin (Ig) isotype switching is a recombination event that changes the constant domain of antibody genes and is catalyzed by activation-induced cytidine deaminase (AID). Upon recruitment to Ig genes, AID deaminates cytidines at switch (S) recombination sites, leading to the formation of DNA breaks. In addition to their role in isotype switching, AID-induced lesions promote Igh-cMyc chromosomal translocations and tumor development. However, cMyc translocations are also present in lymphocytes from healthy humans and mice, and thus, it remains unclear whether AID directly contributes to the dynamics of B cell transformation. Using a plasmacytoma mouse model, we show that AID(+/-) mice have reduced AID expression levels and display haploinsufficiency both in the context of isotype switching and plasmacytomagenesis. At the Ig loci, AID(+/-) lymphocytes show impaired intra- and inter-switch recombination, and a substantial decrease in the frequency of S mutations and chromosomal breaks. In AID(+/-) mice, these defects correlate with a marked decrease in the accumulation of B cell clones carrying Igh-cMyc translocations during tumor latency. These results thus provide a causality link between the extent of AID enzymatic activity, the number of emerging Igh-cMyc-translocated cells, and the incidence of B cell transformation.
Figures
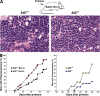
Figure 1.
Happloinsufficiency in plasma cell tumor development in AID+/− mice. (A) Photomicrographs of plasma cell tumors arising in BALB/c-Bcl-xL AID+/+ and BALB/c-Bcl-xL AID+/− mice show no significant variations in plasma cell morphology. Bar, 20 μm. (B, left) Incidence of plasma cell tumors in the presence of Bcl-xL as determined by the histological appearance of foci, with each containing 50 or more atypical plasma cells (red squares, plasma cell tumors in 36 out of 41 AID+/+ mice injected with pristane; black circles, plasma cell tumors in 58 out of 96 AID+/− mice). (right) Plasmacytoma development in AID+/+ (n = 16) and AID+/− (n = 39) mice in the absence of Bcl-xL.
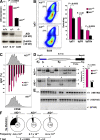
Figure 2.
Reduced AID activity in AID+/− B cells. (A) AID mRNA (graph, AID+/+ and AID+/−; n = 5) and protein (AID+/+, AID+/−, and AID−/−), as measured by real-time PCR and Western blotting. Error bars represent the mean ± SD. (B, left) IgG1 switching (pseudocolor plot) profiles in wild-type and AID+/− B cells activated ex vivo, as determined by flow cytometry. (right) Mean switching to IgG1, IgG3, and IgG2a. Values represent the mean ± SD (n = 5). Antibodies were B220-PercP-Cy5.5 and IgG1-PE. Dead cells were gated out using DAPI. (C) Flow cytometric measurement of cell proliferation and IgG1 switching on CFSE-labeled B cells activated for 96 h with LPS + IL-4. The graph represents percentages of γ1 switching at each cell division (n = 2). Error bars represent the mean ± SD. (D) The percentage of intra-switch deletions as determined by Southern blotting on hybridomas generated from AID+/+ and AID+/− lymphocytes activated for 96 h in the presence of LPS + IL-4. Intact Sμ domains result in a 6.9-kb band, whereas smaller bands (stars) represent intra-switch deletions. (E) Analysis of AID gene expression by single-cell RT-PCR (137 bp) from sorted LPS + IL-4–activated B cells. (F) Sμ mutation frequency in AID+/+, AID+/−, and AID−/− activated B cells. Pie chart segments are proportional to the number of sequences carrying an equal number of mutations (indicated on the periphery). The total number of sequences is portrayed in the middle of each chart, and mutation frequency (calculated as the total mutations per total base pairs sequenced) and p-values are shown below the charts.
Figure 3.
Reduced AID activity in AID+/− mice during the immune response. (A) Measurement of IgG1 levels in GCs from AID+/+ and AID+/− mice. Values represent the mean ± SD (n = 5). (B) Somatic hypermutation as determined by sequencing the JH4 intron from AID+/+, AID+/−, and AID−/− GC cells. (C) ELISA analysis of serum from five AID+/+ and AID+/− mice immunized with NP-CGG. IgG1 antibodies displaying low affinity for NP were captured in this assay using NP30-BSA, whereas high affinity antibodies were monitored with NP7-BSA, as described in Results and discussion. Horizontal bars represent the mean.
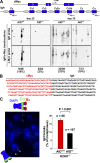
Figure 4.
Paucity of translocation-bearing clones and CSR-induced DNA breaks in AID+/− mice. (A) Igh-cMyc chromosomal translocations in BALB/c-Bcl-xL AID+/+ and BALB/c-Bcl-xL AID+/− mice (three mice per time point) were amplified by PCR 25 or 35 d after pristane injection using nested primers (arrows) specific for cMyc intron 1 and Igh Cμ, Cγ2b, and Cα constant domains. PCR products were analyzed by Southern blotting using _cMyc_- and _Igh_-specific probes. (B) PCR products appearing multiple times were sequenced and counted once if they represented a single clone. (C) Igh FISH in metaphase spreads from AID+/+H2AX−/− and AID+/−H2AX−/− B cells activated for CSR. (left) The loss of the red VH signal with an intact green Cα signal (micrograph) denotes a chromosome 12 break. (right) Total number of breaks observed for each strain (metaphases analyzed are indicated on top of each bar).
Similar articles
- Role of the translocation partner in protection against AID-dependent chromosomal translocations.
Jankovic M, Robbiani DF, Dorsett Y, Eisenreich T, Xu Y, Tarakhovsky A, Nussenzweig A, Nussenzweig MC. Jankovic M, et al. Proc Natl Acad Sci U S A. 2010 Jan 5;107(1):187-92. doi: 10.1073/pnas.0908946107. Epub 2009 Dec 4. Proc Natl Acad Sci U S A. 2010. PMID: 19966290 Free PMC article. - AID is required for c-myc/IgH chromosome translocations in vivo.
Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. Ramiro AR, et al. Cell. 2004 Aug 20;118(4):431-8. doi: 10.1016/j.cell.2004.08.006. Cell. 2004. PMID: 15315756 - Activation-induced cytidine deaminase (AID) promotes B cell lymphomagenesis in Emu-cmyc transgenic mice.
Kotani A, Kakazu N, Tsuruyama T, Okazaki IM, Muramatsu M, Kinoshita K, Nagaoka H, Yabe D, Honjo T. Kotani A, et al. Proc Natl Acad Sci U S A. 2007 Jan 30;104(5):1616-20. doi: 10.1073/pnas.0610732104. Epub 2007 Jan 24. Proc Natl Acad Sci U S A. 2007. PMID: 17251349 Free PMC article. - AID and Igh switch region-Myc chromosomal translocations.
Unniraman S, Schatz DG. Unniraman S, et al. DNA Repair (Amst). 2006 Sep 8;5(9-10):1259-64. doi: 10.1016/j.dnarep.2006.05.019. Epub 2006 Jun 19. DNA Repair (Amst). 2006. PMID: 16784901 Review. - Restricting activation-induced cytidine deaminase tumorigenic activity in B lymphocytes.
Casellas R, Yamane A, Kovalchuk AL, Potter M. Casellas R, et al. Immunology. 2009 Mar;126(3):316-28. doi: 10.1111/j.1365-2567.2008.03050.x. Immunology. 2009. PMID: 19302140 Free PMC article. Review.
Cited by
- Activation-induced cytidine deaminase in antibody diversification and chromosome translocation.
Gazumyan A, Bothmer A, Klein IA, Nussenzweig MC, McBride KM. Gazumyan A, et al. Adv Cancer Res. 2012;113:167-90. doi: 10.1016/B978-0-12-394280-7.00005-1. Adv Cancer Res. 2012. PMID: 22429855 Free PMC article. Review. - Msh6 protects mature B cells from lymphoma by preserving genomic stability.
Peled JU, Sellers RS, Iglesias-Ussel MD, Shin DM, Montagna C, Zhao C, Li Z, Edelmann W, Morse HC 3rd, Scharff MD. Peled JU, et al. Am J Pathol. 2010 Nov;177(5):2597-608. doi: 10.2353/ajpath.2010.100234. Epub 2010 Oct 7. Am J Pathol. 2010. PMID: 20934970 Free PMC article. - AID downregulation is a novel function of the DNMT inhibitor 5-aza-deoxycytidine.
Tsai CT, Yang PM, Chern TR, Chuang SH, Lin JH, Klemm L, Müschen M, Chen CC. Tsai CT, et al. Oncotarget. 2014 Jan 15;5(1):211-23. doi: 10.18632/oncotarget.1319. Oncotarget. 2014. PMID: 24457556 Free PMC article. - Mechanisms of Yin Yang 1 in oncogenesis: the importance of indirect effects.
Atchison M, Basu A, Zaprazna K, Papasani M. Atchison M, et al. Crit Rev Oncog. 2011;16(3-4):143-61. doi: 10.1615/critrevoncog.v16.i3-4.20. Crit Rev Oncog. 2011. PMID: 22248052 Free PMC article. Review. - An efficient method to enrich for knock-out and knock-in cellular clones using the CRISPR/Cas9 system.
Niccheri F, Pecori R, Conticello SG. Niccheri F, et al. Cell Mol Life Sci. 2017 Sep;74(18):3413-3423. doi: 10.1007/s00018-017-2524-y. Epub 2017 Apr 18. Cell Mol Life Sci. 2017. PMID: 28421278 Free PMC article.
References
- Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. - PubMed
- Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575. - PubMed
- Di Noia, J., and M.S. Neuberger. 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 419:43–48. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases