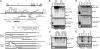An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor - PubMed (original) (raw)
An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor
Shuang Tang et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Latency-associated transcript (LAT) sequences regulate herpes simplex virus (HSV) latency and reactivation from sensory neurons. We found a HSV-2 LAT-related microRNA (miRNA) designated miR-I in transfected and infected cells in vitro and in acutely and latently infected ganglia of guinea pigs in vivo. miR-I is also expressed in human sacral dorsal root ganglia latently infected with HSV-2. miR-I is expressed under the LAT promoter in vivo in infected sensory ganglia. We also predicted and identified a HSV-1 LAT exon-2 viral miRNA in a location similar to miR-I, implying a conserved mechanism in these closely related viruses. In transfected and infected cells, miR-I reduces expression of ICP34.5, a key viral neurovirulence factor. We hypothesize that miR-I may modulate the outcome of viral infection in the peripheral nervous system by functioning as a molecular switch for ICP34.5 expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
HSV-2 LAT region encodes a miRNA. (A) Schematic diagram of HSV-2 LAT region and HSV-2 miR-I. Restriction endonucleases used to create mutant viruses and plasmids are labeled. Stable RNAs including primary LAT, the LAT intron, ICP0, ICP34.5, ICP4, ORF-O (putative), and ORF-P (putative) are labeled based on their relative transcription-starting sites and transcription direction. The location of a TATA box in the LAT intron, which is mutated in the ΔYAT virus, is also labeled. The HSV-2 mature miRNA (bold) was identified by small-RNA cloning and maps to HSV-2 LAT exon 2 (nucleotides 569–547 and 126681–126703). The predicted anti-sense strand of miR-I is shown in italics. ≈50% of miR-I sequences cloned had one additional “C” at their 3′ end. The arrows indicate Dicer cutting sites. Mutant HSV-2 viruses and HSV-2 LAT plasmids are also shown. TRL, terminal repeat long; IRL, internal repeat long; IRS, internal repeat short; US, unique short; TRS, terminal repeat short; UL, unique long. The open boxes on ICP0 and ICP34.5 represent introns. (B) HSV-2 miR-I detection by Northern blot in 293 and HeLa cells transfected with plasmids containing full-length LAT gene but not with truncated LAT plasmids. Total RNAs from HEK 293 cells and HeLa cells transfected with or without plasmids pSSK and pCMV-SSK, which include the ICP34.5 coding region, and plasmids pSSB and pCMV-SSB, which are truncated and lack the ICP34.5 region of LAT, were blotted with 32P-labeled oligo probe for miR-I. The same membrane was stripped and reprobed with an oligonucleotide probe for the predicted anti-sense strand of miR-I and a probe for U6 small nuclear RNA (snRNA). (C) Mature miR-I was significantly reduced, but pre-miRNA increased in Dicer exon-5 disrupted cells. Wild-type (WT) and Dicer exon-5 disrupted cells (Dicer−/−) were studied with or without HSV-2 strain HG52 infection. Total RNAs were hybridized with the HSV-2 miR-I probe and the U6 probe. (D) LAT promoter is not the sole promoter for HSV-2 miR-I production in infected cell cultures. Vero cells were infected with HSV-2 strain HG52, HSV-2 strain 333, and HSV-2 mutant viruses including ΔLAT, CMV-LAT, ΔYAT, and ΔNot-SalI at 0.1 multiplicity of infection or mock-infected. miR-I, the anti-sense strand of miR-I, and U6 snRNA were detected by Northern hybridization after stripping the same membrane. (E) The sequences directly upstream of miR-I contribute to miR-I expression but to a lesser extent than LAT promoter sequences in transfected cells. HEK 293 cells and HeLa cells were transfected with pSSK and pPstI-HincII, which does not contain the LAT promoter region but contains ≈3 kb of sequence upstream of miR-I, or mock-transfected. Total RNA from these transfected cells were hybridized with HSV-2 miR-I and U6 snRNA probes.
Fig. 2.
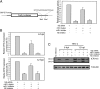
HSV-2 miR-I is highly expressed in guinea pig ganglia during latency, and the LAT promoter accounts for production of miR-I in both acutely and latently infected dorsal root ganglia. Total RNA was prepared from dorsal root ganglia of guinea pigs infected with DLAT acutely (n = 3) and latently (n = 2), HSV-2 ΔR (ΔLAT rescuant virus) acutely (n = 3) and latently (n = 3), or with HSV-1 17syn+ acutely (n = 3) and latently (n = 3). miR-I-specific real-time PCR was used to detect miR-I. A synthetic single-stranded miR-I was used as a standard. All animals acutely or latently infected with HSV-2 ΔR showed high levels of miR-I expression. One animal acutely and one animal latently infected with ΔLAT gave very low miR-I signals, and the others were under the detectable level. miR-I copy numbers in different groups are indicated in panel (A). The copy numbers of LAT RNA, virus DNA, and miR-I are shown in panel (B). HSV-1 viral DNA and LAT copies were measured with HSV-1 specific primers and _Taq_man probes.
Fig. 3.
miR-I can efficiently silence target gene expression. (A) HSV-2 miR-I efficiently silences the expression of a firefly luciferase reporter with miR-I target sequence in its 3′ UTR. (Left) Shown is a diagram of the HSV-2 miR-I reporter. (Right) HEK 293 cells were cotransfected with the firefly luciferase reporter plasmid with miR-I target sequence, Renilla luciferase plasmid, and either 2 nM NS-siRNA control or 2 nM miR-I duplex, 2 nM miR-I plus 30 nM miR-I inhibitor, or NS inhibitor. Relative luciferase activity was defined as the ratio of firefly luciferase activity to Renilla luciferase activity. miR-I, but not the nonspecific siRNA control (NS-siRNA), efficiently silenced the target reporter expression, whereas the activity of cotransfected Renilla luciferase was unaffected by miR-I. miR-I-specific inhibitor, but not NS inhibitor, rescued the inhibition of luciferase reporter activity by miR-I. The figure is a summary of four independent experiments. (B) miR-I efficiently knocks down ICP34.5 expression in infected cell culture detected by real-time PCR. Forty nanomolar of miR-I duplex or nonspecific NS-siRNA was transfected with or without 100 nM miR-I inhibitor or NS inhibitor into U2OS cells 18 h before infection with HSV-2 strain 333. After 6 and 12 hpi, total RNAs were extracted, and the uncut ICP34.5, and thymidine kinase (TK) were detected by real-time PCR. Relative ICP34.5 expression was defined as the ratio of ICP34.5 to TK. The figure represents a summary of four independent experiments. (C) miR-I efficiently knocks down ICP34.5 protein expression in infected cell culture. Twenty nanomolar miR-I duplex or nonspecific NS-siRNA was transfected with or without 100 nM miR-I inhibitor or NS inhibitor as described B, and HSV-2 ICP34.5 was detected by Western blot. The same membrane was stripped and reblotted with an anti-β-tubulin antibody as the internal loading control.
Fig. 4.
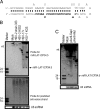
HSV-1 also encodes a miRNA in LAT exon 2, overlapping the anti-sense strand of the ICP34.5 gene. (A) Diagram of HSV-1 miR-LAT-ICP34.5. miR-LAT-ICP34.5 sequence (nucleotides 429–449 and 125942–125922) is shown in bold italics. (B) Detection of HSV-1 miR-LAT-ICP34.5 in HSV-1-infected cells. HSV-1 miR-LAT-ICP34.5 was detected by Northern blot in cells infected with either HSV-1 strains 17syn+ or KOS but not HSV-2 strains. The anti-sense strand of miR-LAT-ICP34.5 was not detected when hybridized with anti-sense strand probe. (C) Detection of miR-LAT-ICP34.5 in cells transfected with HSV-1 LAT plasmids. miR-LAT-ICP34.5 was detected in 293 cells transfected with pAvrII-SapI, which contains the entire HSV-1 LAT promoter and LAT sequences, but not in cells transfected with pAvrII-AluI, which is truncated and lacks miR-LAT-ICP34.5 sequences.
Similar articles
- Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs.
Tang S, Patel A, Krause PR. Tang S, et al. J Virol. 2009 Feb;83(3):1433-42. doi: 10.1128/JVI.01723-08. Epub 2008 Nov 19. J Virol. 2009. PMID: 19019961 Free PMC article. - Characterization of herpes simplex virus 2 primary microRNA Transcript regulation.
Tang S, Bosch-Marce M, Patel A, Margolis TP, Krause PR. Tang S, et al. J Virol. 2015 May;89(9):4837-48. doi: 10.1128/JVI.03135-14. Epub 2015 Feb 11. J Virol. 2015. PMID: 25673716 Free PMC article. - A comparison of herpes simplex virus type 1 and varicella-zoster virus latency and reactivation.
Kennedy PG, Rovnak J, Badani H, Cohrs RJ. Kennedy PG, et al. J Gen Virol. 2015 Jul;96(Pt 7):1581-602. doi: 10.1099/vir.0.000128. Epub 2015 Mar 20. J Gen Virol. 2015. PMID: 25794504 Free PMC article. Review. - The latency-associated gene of herpes simplex virus type 1 (HSV-1) interferes with superinfection by HSV-1.
Mador N, Panet A, Steiner I. Mador N, et al. J Neurovirol. 2002 Dec;8 Suppl 2:97-102. doi: 10.1080/13550280290167920. J Neurovirol. 2002. PMID: 12491159 Review.
Cited by
- LAT region factors mediating differential neuronal tropism of HSV-1 and HSV-2 do not act in trans.
Bertke AS, Apakupakul K, Ma A, Imai Y, Gussow AM, Wang K, Cohen JI, Bloom DC, Margolis TP. Bertke AS, et al. PLoS One. 2012;7(12):e53281. doi: 10.1371/journal.pone.0053281. Epub 2012 Dec 31. PLoS One. 2012. PMID: 23300908 Free PMC article. - The HSV-1 Latency-Associated Transcript Functions to Repress Latent Phase Lytic Gene Expression and Suppress Virus Reactivation from Latently Infected Neurons.
Nicoll MP, Hann W, Shivkumar M, Harman LE, Connor V, Coleman HM, Proença JT, Efstathiou S. Nicoll MP, et al. PLoS Pathog. 2016 Apr 7;12(4):e1005539. doi: 10.1371/journal.ppat.1005539. eCollection 2016 Apr. PLoS Pathog. 2016. PMID: 27055281 Free PMC article. - Impairment in reactivation of a latency associated transcript (LAT)-deficient HSV-2 is not solely dependent on the latent viral load or the number of CD8(+) T cells infiltrating the ganglia.
Hoshino Y, Pesnicak L, Straus SE, Cohen JI. Hoshino Y, et al. Virology. 2009 Apr 25;387(1):193-9. doi: 10.1016/j.virol.2009.02.004. Epub 2009 Mar 9. Virology. 2009. PMID: 19269661 Free PMC article. - Herpes Simplex Virus 1 Lytic Infection Blocks MicroRNA (miRNA) Biogenesis at the Stage of Nuclear Export of Pre-miRNAs.
Pan D, Li G, Morris-Love J, Qi S, Feng L, Mertens ME, Jurak I, Knipe DM, Coen DM. Pan D, et al. mBio. 2019 Feb 12;10(1):e02856-18. doi: 10.1128/mBio.02856-18. mBio. 2019. PMID: 30755517 Free PMC article. - Investigation of the mechanism by which herpes simplex virus type 1 LAT sequences modulate preferential establishment of latent infection in mouse trigeminal ganglia.
Imai Y, Apakupakul K, Krause PR, Halford WP, Margolis TP. Imai Y, et al. J Virol. 2009 Aug;83(16):7873-82. doi: 10.1128/JVI.00043-09. Epub 2009 Jun 3. J Virol. 2009. PMID: 19493993 Free PMC article.
References
- Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials