The multiple lives of NMD factors: balancing roles in gene and genome regulation - PubMed (original) (raw)
Review
The multiple lives of NMD factors: balancing roles in gene and genome regulation
Olaf Isken et al. Nat Rev Genet. 2008 Sep.
Abstract
Nonsense-mediated mRNA decay (NMD) largely functions to ensure the quality of gene expression. However, NMD is also crucial to regulating appropriate expression levels for certain genes and for maintaining genome stability. Furthermore, just as NMD serves cells in multiple ways, so do its constituent proteins. Recent studies have clarified that UPF and SMG proteins, which were originally discovered to function in NMD, also have roles in other pathways, including specialized pathways of mRNA decay, DNA synthesis and cell-cycle progression, and the maintenance of telomeres. These findings suggest a delicate balance of metabolic events - some not obviously related to NMD - that can be influenced by the cellular abundance, location and activity of NMD factors and their binding partners.
Figures
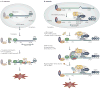
Figure 1. Models for nonsense-mediated mRNA decay in Saccharomyces cerevisiae and mammals
a | In Saccharomyces cerevisiae, newly synthesized mRNAs that contain a premature termination codon (PTC) and that are bound to the RNA cap-binding protein heterodimer Cbc1–Cbc2 and to steady-state mRNAs that are bound by the cap-binding protein eukaryotic translation initiation factor 4E (eIF4E) are targeted for nonsense-mediated mRNA decay (NMD) once the mRNA is exported from the nucleus to the cytoplasm. In at least one mechanism, an abnormally long or ‘faux’ 3′ UTR results in inefficient translation termination. As a consequence, termination involves not only the eukaryotic release factor 1 (eRF1) and eRF3 translation-termination factors, which fail to effectively mediate the release of the nascent polypeptide because of an inefficient interaction between eRF3 and poly(A)-binding protein 1 (Pab1), but probably also the up-frameshift 1 (Upf1), Upf2 and Upf3 NMD factors. These factors then recruit and/or activate mRNA degradative activities. Although Upf1 is a phosphoprotein, whether Upf1 undergoes a cycle of phosphorylation and dephosphorylation during NMD in S. cerevisiae is unknown. b | In mammals, newly synthesized PTC-containing mRNA that is bound to the RNA cap-binding protein heterodimer CPB80–CBP20 is targeted for NMD once the mRNA has been generated by pre-mRNA splicing and exported from the nucleus to the cytoplasm. Notably, pre-mRNA splicing results in the deposition of an exon-junction complex (EJC) of proteins upstream of mRNA exon–exon junctions. Core EJC components consist of eIF4AIII, RNA-binding-motif protein Y14, mago nashi homologue (MAGOH) and Barentsz (BTZ; also known as cancer susceptibility candidate 3, CASC3). The UPF3 or UPF3X NMD factor, which shuttles to the nucleus, is thought to be recruited to EJCs in the nucleus and is exported with the mRNA to the cytoplasm. UPF3 or UPF3X then recruits UPF2, which is primarily cytoplasmic. The translation of mRNA that is bound to CBP80–CBP20 is termed the pioneer round. Translation termination at a PTC during the pioneer round involves the SURF complex, which consists of the phosphatidylinositol 3-kinase-related protein kinase (PIKK) SMG1 together with UPF1, eRF1 and eRF3. Generally, if translation terminates more than ~50–55 nucleotides (nt) upstream of an exon–exon junction (that is, more than ~25–30 nt upstream of an EJC), then NMD will occur. UPF1, together with SMG1, is thought to bind EJC-associated UPF2 in a way that is promoted by CBP80 (not shown). UPF1 binding to the EJC triggers UPF1 phosphorylation and NMD by promoting translational repression and recruiting mRNA degradative activities. Not shown are SMG5, SMG6 and SMG7, which seem to recruit protein phosphatase 2A (PP2A) and function in UPF1 dephosphorylation and, thus, recycling. AUG, translation initiation codon; STOP, normal termination codon.
Figure 2. UPF1 function in specialized mRNA-decay pathways
a | Staufen 1 (STAU1)-mediated mRNA decay targets particular newly synthesized mRNAs that contain a STAU1 binding site (SBS) within their 3′ UTR and that are bound to the cap-binding protein heterodimer CBP80–CBP20 and to the corresponding steady-state mRNAs that are bound to the cap-binding protein eukaryotic translation initiation factor 4E (eIF4E). SBS-bound STAU1 recruits the NMD factor up-frameshift 1 (UPF1), which presumably undergoes phosphorylation by a protein kinase (yet to be identified) when translation terminates more than ~20–25 nucleotides (nt) upstream of the SBS. b | Replication-dependent histone mRNAs are degraded at the end of S phase of the cell cycle or following replication stress. These histone mRNAs contain a binding site for the stem-loop binding protein (SLBP) within their 3′ UTR. 3′-UTR-bound SLBP recruits UPF1, which seems to undergo phosphorylation mediated by ataxia-telangiectasia mutated and Rad 3-related (ATR) or DNA-dependent protein kinase (DNA-PK) when translation terminates anywhere in the 22–77 nucleotide region upstream of the SLBP-binding site. The 77-nucleotide limit is because of the need for SLBP to be situated sufficiently close to the termination codon to function in translation termination. Notably, it is primarily eIF4E-bound histone mRNA that is targeted for decay at the end of S phase owing to the concomitant downregulation of histone gene transcription. AUG, translation initiation codon; eRF, eukaryotic release factor; STOP, normal translation termination codon: PABP1, poly(A)-binding protein 1.
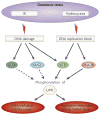
Figure 3. Phosphorylated UPF1 functions when DNA is damaged or replication is otherwise blocked
Exposure of cells to different types of genotoxic stress such as ionizing radiation (IR) or hydroxyurea induces DNA damage and/or a block in DNA replication. These abnormalities activate one or more of the phosphatidylinositol 3-kinase-related protein kinase (PIKK) signal transducers ataxia-telangiectasia mutated (ATM), ataxia-telangiectasia mutated and Rad 3-related (ATR), SMG1 or DNA-dependent protein kinase (DNA-PK), which stimulate phosphorylation of target proteins, such as the NMD factor up-frameshift 1 (UPF1). The resulting phosphorylated UPF1 then functions in DNA replication and repair as well as in histone mRNA decay. The multiple roles of UPF1 in these processes, all presumably by its phosphorylation by PIKKs, ensures that histone production is closely coupled to the cellular need for newly synthesized chromatin.
Figure 4. UPF1 and SMG1 are at the interface of processes important for gene and genome regulation in mammalian cells
(Anticlockwise from the upper left). Up-frameshift 1 (UPF1) and phosphatidylinositol 3-kinase-related protein kinase SMG1 are involved in multiple RNA and DNA surveillance pathways. UPF1 functions during nonsense-mediated mRNA decay (NMD), in which translation termination that occurs sufficiently upstream of a splicing-generated exon-junction complex7 (EJC), usually at a premature termination codon (PTC), results in SMG1-mediated UPF1 phosphorylation and, as a consequence, mRNA decay. UPF1 is also instrumental to the RNA-binding protein Staufen 1 (STAU1)-mediated mRNA decay (SMD) and replication-dependent histone mRNA decay, during which it is recruited to specific 3′ UTRs by STAU1 (in SMD) or stem-loop binding protein (SLBP) (in replication-dependent histone mRNA decay). Although the UPF1 kinase involved in SMD is unknown, ataxia-telangiectasia mutated and Rad 3-related (ATR), ataxia-telangiectasia mutated (ATM) and DNA-dependent protein kinase (DNA-PK) seem to phosphorylate UPF1 during histone mRNA decay. Additionally, UPF1 associates with DNA polymerase (Pol) δ and is essential for human cells to complete DNA replication and repair in a process that involves ATR if not other phosphatidylinositol 3-kinase-related protein kinases (PIKKs). Finally, UPF and SMG proteins seem to function in genome stability to modulate telomerase function and to regulate telomere length: they are enriched at telomeres so as to negatively regulate telomeric repeat-containing RNA (TERRA) association with telomeric chromosomes. AUG, translation initiation codon; BTZ, Barentsz (also known as cancer susceptibility candidate 3, CASC3); eIF4AIII, eukaryotic initiation factor 4AIII; MAGOH, mago nashi homologue; PTC, premature translation termination codon; SBS, STAU1 binding site; STOP, normal termination codon; Y14, RNA-binding motif-protein Y14.
Similar articles
- NMD mechanism and the functions of Upf proteins in plant.
Dai Y, Li W, An L. Dai Y, et al. Plant Cell Rep. 2016 Jan;35(1):5-15. doi: 10.1007/s00299-015-1867-9. Epub 2015 Sep 23. Plant Cell Rep. 2016. PMID: 26400685 Review. - Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution.
Maderazo AB, Belk JP, He F, Jacobson A. Maderazo AB, et al. Mol Cell Biol. 2003 Feb;23(3):842-51. doi: 10.1128/MCB.23.3.842-851.2003. Mol Cell Biol. 2003. PMID: 12529390 Free PMC article. - Upf proteins: highly conserved factors involved in nonsense mRNA mediated decay.
Gupta P, Li YR. Gupta P, et al. Mol Biol Rep. 2018 Feb;45(1):39-55. doi: 10.1007/s11033-017-4139-7. Epub 2017 Dec 27. Mol Biol Rep. 2018. PMID: 29282598 Review. - Chapter 9. Studying nonsense-mediated mRNA decay in mammalian cells.
Matsuda D, Sato H, Maquat LE. Matsuda D, et al. Methods Enzymol. 2008;449:177-201. doi: 10.1016/S0076-6879(08)02409-9. Methods Enzymol. 2008. PMID: 19215759 - mRNAs encoding telomerase components and regulators are controlled by UPF genes in Saccharomyces cerevisiae.
Dahlseid JN, Lew-Smith J, Lelivelt MJ, Enomoto S, Ford A, Desruisseaux M, McClellan M, Lue N, Culbertson MR, Berman J. Dahlseid JN, et al. Eukaryot Cell. 2003 Feb;2(1):134-42. doi: 10.1128/EC.2.1.134-142.2003. Eukaryot Cell. 2003. PMID: 12582130 Free PMC article.
Cited by
- Whole-exome sequencing, without prior linkage, identifies a mutation in LAMB3 as a cause of dominant hypoplastic amelogenesis imperfecta.
Poulter JA, El-Sayed W, Shore RC, Kirkham J, Inglehearn CF, Mighell AJ. Poulter JA, et al. Eur J Hum Genet. 2014 Jan;22(1):132-5. doi: 10.1038/ejhg.2013.76. Epub 2013 May 1. Eur J Hum Genet. 2014. PMID: 23632796 Free PMC article. - Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity.
Park E, Gleghorn ML, Maquat LE. Park E, et al. Proc Natl Acad Sci U S A. 2013 Jan 8;110(2):405-12. doi: 10.1073/pnas.1213508110. Epub 2012 Dec 20. Proc Natl Acad Sci U S A. 2013. PMID: 23263869 Free PMC article. - Mutually exclusive splicing regulates the Nav 1.6 sodium channel function through a combinatorial mechanism that involves three distinct splicing regulatory elements and their ligands.
Zubović L, Baralle M, Baralle FE. Zubović L, et al. Nucleic Acids Res. 2012 Jul;40(13):6255-69. doi: 10.1093/nar/gks249. Epub 2012 Mar 19. Nucleic Acids Res. 2012. PMID: 22434879 Free PMC article. Retracted. - Posttranscriptional regulation by RNA-binding proteins during epithelial-to-mesenchymal transition.
Aparicio LA, Abella V, Valladares M, Figueroa A. Aparicio LA, et al. Cell Mol Life Sci. 2013 Dec;70(23):4463-77. doi: 10.1007/s00018-013-1379-0. Epub 2013 May 29. Cell Mol Life Sci. 2013. PMID: 23715860 Free PMC article. Review. - The plausible reason why the length of 5' untranslated region is unrelated to organismal complexity.
Chen CH, Lin HY, Pan CL, Chen FC. Chen CH, et al. BMC Res Notes. 2011 Aug 27;4:312. doi: 10.1186/1756-0500-4-312. BMC Res Notes. 2011. PMID: 21871111 Free PMC article.
References
- Behm-Ansmant I, et al. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. - PubMed
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. - PubMed
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. A comprehensive review of translation-dependent mRNA surveillance mechanisms, with a focus on NMD. - PubMed
- Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nature Genet. 2004;36:801–808. - PubMed
- Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nature Rev Mol Cell Biol. 2006;7:415–425. A comprehensive overview of NMD in S. cerevisiae. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources