Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism - PubMed (original) (raw)
Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism
Siming Li et al. Cell Metab. 2008 Aug.
Abstract
Impaired mitochondrial function has been implicated in the pathogenesis of type 2 diabetes, heart failure, and neurodegeneration as well as during aging. Studies with the PGC-1 transcriptional coactivators have demonstrated that these factors are central components of the regulatory network that controls mitochondrial function in mammalian cells. Here we describe a genome-wide coactivation assay to globally identify transcription factors and cofactors in this pathway. These analyses revealed a molecular signature of the PGC-1alpha transcriptional network and identified BAF60a (SMARCD1) as a molecular link between the SWI/SNF chromatin-remodeling complexes and hepatic lipid metabolism. Adenoviral-mediated expression of BAF60a stimulates fatty acid beta-oxidation in cultured hepatocytes and ameliorates hepatic steatosis in vivo. PGC-1alpha mediates the recruitment of BAF60a to PPARalpha-binding sites, leading to transcriptional activation of peroxisomal and mitochondrial fat-oxidation genes. These results define a role for the SWI/SNF complexes in the regulation of lipid homeostasis.
Figures
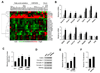
Figure 1. Identification of Transcriptional Partners for PGC-1α Through Genome-wide Coactivation Screen
(A) Schematic diagram of high-throughput coactivation screen of PGC-1α. (B) Log-transformed fold-coactivation by PGC-1α for individual Gal-TF clones. Red line denotes 10-fold increase in luciferase activity by PGC-1α. (C) Representative graphs of CoIP between PGC-1α and 11 putative TF partners. Immunoblotting was performed on total lysates and immunoprecipitated complexes from BOSC cells transiently transfected with Myc-TF with pcDNA3 vector or Flag-PGC-1α. (D) Schematic diagram of the PGC-1α transcriptional regulatory network. Previously reported (green line) and new factors identified in this study (red line) are indicated.
Figure 2. Regulation of Fatty Acid β-oxidation by BAF60a in Primary Hepatocytes
(A) Clustering analysis of gene expression in primary hepatocytes transduced with adenoviruses expressing GFP, PGC-1α, or individual TFs. Relative mRNA expression of genes involved in hepatic gluconeogenesis, fatty acid β-oxidation, mitochondrial OXPHOS, and circadian clock were analyzed using qPCR. (B) qPCR analysis of gene expression in hepatocytes transduced with GFP (open) or BAF60a adenoviruses at multiplicity of infection of 1 (grey) or 3 (filled). (C) qPCR analysis of relative mitochondrial/nuclear DNA content. (D) Immunoblotting analysis of transduced primary hepatocytes. (E–F) Relative rate of fatty acid oxidation (E) and secretion of TG-containing lipoproteins (F) in transduced hepatocytes. All data represent mean ± stdev. *p<0.05.
Figure 3. Adenoviral-mediated Expression of BAF60a Lowers Hepatic TG Content
(A) Immunoblots of liver nuclear extracts from transduced mice using indicated antibodies. The expression of endogenous (En-BAF60a) and adenoviral (Ad-BAF60a) BAF60a proteins is indicated. (B) Liver morphology and histological staining of liver sections from mice six days after tail vein injection. (C) Liver TG, plasma lipid and glucose concentrations in mice transduced with GFP or BAF60a adenoviruses. (D) Increase in plasma TG following intravenous injection of tyloxapol (500 mg/kg) in mice transduced with GFP (open circle) or BAF60a (filled circle) adenoviruses. (E) qPCR analysis of hepatic gene expression in mice transduced with GFP (open) or BAF60a (filled) adenoviruses. Data in C–E represent mean ± SEM, n=5. *p<0.05.
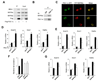
Figure 4. Physical and Functional Interaction Between BAF60a and PGC-1α
(A) Immunoblots of total lysates or immunoprecipitated proteins from BOSC cells transiently transfected with pcDNA3 vector or Flag-PGC-1α plasmids. Note that PGC-1α associates with endogenous BAF60a, BRG-1, and BAF53a. (B) Immunoblots of precipitated liver nuclear complexes using IgG and PGC-1α antibodies. (C) Confocal microscopy images of H2.35 hepatoma cells transiently transfected with GFP-PGC-1α and RFP-BAF60a plasmids. (D) qPCR analysis of gene expression in primary hepatocytes transduced with adenoviruses expressing GFP, BAF60a, PGC-1α, or the combination of BAF60a and PGC-1α. Data represent mean ± stdev. *p<0.01. (E) qPCR analysis of gene expression in wild type (filled) and PGC-1α null (open) hepatocytes transduced with GFP or BAF60a adenoviruses, as indicated. (F) FAO rate in transduced hepatocytes isolated from wild type (filled) and PGC-1α null (open) mice. (G) qPCR analysis of gene expression in wild type (filled) and PGC-1α null (open) mouse livers transduced with GFP or BAF60a adenoviruses. Data in (E–G) represent mean ± stdev. *p<0.05 WT vs. PGC-1α null.
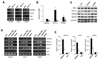
Figure 5. Crosstalk Between PPARα and BAF60a in the Regulation of Hepatic Fat Oxidation
(A) qPCR analysis of gene expression in hepatocytes transduced with GFP or BAF60a adenoviruses followed by vehicle (−) or 10 µM Wy14,643 (+) treatments. Data represent mean ± stdev. *p<0.05 Wy14,643 vs. vehicle. (B) Venn diagram representation of FAO genes that are induced more than 1.6-fold by BAF60a or Wy14,643 treatment in hepatocytes. (C) Immunoblots of precipitated liver nuclear complexes using IgG and PPARα antibodies. (D) Immunoblots of Flag-tagged BAF60a and PGC-1α. GST-PPARα fusion protein was incubated with in vitro transcribed and translated Flag-BAF60a or Flag-PGC-1α in the absence or presence of 20 µM Wy14,643. (E) Immunoblots of Flag-tagged full length (FL) or truncated PPARα proteins. Recombinant GST, GST-PGC-1α (1–400) or GST-BAF60a proteins were incubated with in vitro transcribed and translated PPARα followed by immunoblotting analysis. (F) Immunoblots of Flag-tagged BAF60a and PPARα. GST fusion proteins containing various domains of PGC-1α were incubated with in vitro transcribed and translated Flag-BAF60a or Flag-PPARα followed by immunoblotting analysis. (G) Immunoblots of Flag-tagged PPARα and PGC-1α. GST fusion proteins containing various domains of BAF60a were incubated with in vitro transcribed and translated Flag-PPARα or Flag-PGC-1α followed by immunoblotting analysis. (H) qPCR analysis of gene expression in wild type (filled) or PPARα null (open) hepatocytes transduced with GFP or BAF60a adenoviruses, as indicated. Data represent mean ± stdev. *p<0.05 WT vs. PPARα null. (I) qPCR analysis of gene expression in wild type (filled) or PPARα null (open) mouse livers transduced with GFP or BAF60a adenoviruses. Data represent mean ± stdev. *p<0.05 WT vs. PPARα null.
Figure 6. Recruitment of BAF60a to FAO Genes is Induced by Fasting
(A) ChIP assay using BAF60a or AcH3 antibodies in H2.35 cells transduced with GFP or PGC-1α adenoviruses. PCR primers flanking PPRE present in the promoters of Acaa1b, Acox1, and Hadha were used. (B) qPCR analysis of BAF60a, PGC-1α, and PPARα expression in fed (open), fasted (24 hrs, filled), and fasted/refed (24/20 hrs, grey) mouse livers. Data represent mean ± SEM, n=4. *p<0.05. (C) Immunoblots of liver nuclear extracts using indicated antibodies. (D) ChIP assays with chromatin extracts prepared from fed, fasted and refed mouse livers using indicated antibodies. PCR primers are the same as in (A). (E) ChIP-qPCR assay with chromatin lysates prepared from 24-hr fasted wild type (filled) or PGC-1α null (open) mouse livers using BAF60a antibody. Data represent mean ± stdev, *p<0.01 wild type vs. PGC-1α null.
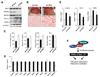
Figure 7. BAF60a is Required for Hepatic Fat Oxidation During Starvation
(A) Immunoblots of liver nuclear extracts from mice transduced with control (Ad-Scrb) or BAF60a RNAi (Ad-RNAi) adenoviruses. (B) Liver morphology and histological staining of liver sections in transduced mice. Mice were subjected to 40-hr fasting before metabolic measurements and gene expression analysis. (C) Hepatic TG, plasma lipid and glucose concentrations in mice transduced with control (filled) or BAF60a RNAi (open) adenoviruses. (D) qPCR analysis of hepatic gene expression in mice transduced with control (filled) or BAF60a RNAi (open) adenoviruses. Data in (C–D) represent mean ± SEM (n=4). *p<0.05. (E) qPCR analysis of hepatic gene expression in mice transduced with control or BAF60a RNAi adenoviruses followed by oral gavage with vehicle (open) or 300 mg Wy14,643 (filled). Data represent mean ± stdev, *p<0.05. (F) Schematic model of the regulation of hepatic FAO genes by BAF60a and its associated SWI/SNF complexes. Note the nutritional regulation of BAF60a recruitment to FAO genes through PPARα and other transcription factors.
Similar articles
- SWI/SNF complex subunit BAF60a represses hepatic ureagenesis through a crosstalk between YB-1 and PGC-1α.
Zhang W, Dong Z, Xu M, Zhang S, Liu C, Chen S. Zhang W, et al. Mol Metab. 2020 Feb;32:85-96. doi: 10.1016/j.molmet.2019.12.007. Epub 2019 Dec 20. Mol Metab. 2020. PMID: 32029232 Free PMC article. - Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway.
Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, Lawrence JC Jr, Kelly DP. Finck BN, et al. Cell Metab. 2006 Sep;4(3):199-210. doi: 10.1016/j.cmet.2006.08.005. Cell Metab. 2006. PMID: 16950137 - Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle.
Huss JM, Torra IP, Staels B, Giguère V, Kelly DP. Huss JM, et al. Mol Cell Biol. 2004 Oct;24(20):9079-91. doi: 10.1128/MCB.24.20.9079-9091.2004. Mol Cell Biol. 2004. PMID: 15456881 Free PMC article. - Peroxisome proliferator-activated receptors, coactivators, and downstream targets.
Qi C, Zhu Y, Reddy JK. Qi C, et al. Cell Biochem Biophys. 2000;32 Spring:187-204. doi: 10.1385/cbb:32:1-3:187. Cell Biochem Biophys. 2000. PMID: 11330046 Review. - Minireview: the PGC-1 coactivator networks: chromatin-remodeling and mitochondrial energy metabolism.
Lin JD. Lin JD. Mol Endocrinol. 2009 Jan;23(1):2-10. doi: 10.1210/me.2008-0344. Epub 2008 Nov 13. Mol Endocrinol. 2009. PMID: 19008463 Free PMC article. Review.
Cited by
- SEC24A deficiency lowers plasma cholesterol through reduced PCSK9 secretion.
Chen XW, Wang H, Bajaj K, Zhang P, Meng ZX, Ma D, Bai Y, Liu HH, Adams E, Baines A, Yu G, Sartor MA, Zhang B, Yi Z, Lin J, Young SG, Schekman R, Ginsburg D. Chen XW, et al. Elife. 2013 Apr 9;2:e00444. doi: 10.7554/eLife.00444. Elife. 2013. PMID: 23580231 Free PMC article. - Neuregulin 4 suppresses NASH-HCC development by restraining tumor-prone liver microenvironment.
Zhang P, Chen Z, Kuang H, Liu T, Zhu J, Zhou L, Wang Q, Xiong X, Meng Z, Qiu X, Jacks R, Liu L, Li S, Lumeng CN, Li Q, Zhou X, Lin JD. Zhang P, et al. Cell Metab. 2022 Sep 6;34(9):1359-1376.e7. doi: 10.1016/j.cmet.2022.07.010. Epub 2022 Aug 15. Cell Metab. 2022. PMID: 35973424 Free PMC article. - The Baf60c/Deptor pathway links skeletal muscle inflammation to glucose homeostasis in obesity.
Meng ZX, Wang L, Xiao Y, Lin JD. Meng ZX, et al. Diabetes. 2014 May;63(5):1533-45. doi: 10.2337/db13-1061. Epub 2014 Jan 23. Diabetes. 2014. PMID: 24458360 Free PMC article. - Otopetrin 1 protects mice from obesity-associated metabolic dysfunction through attenuating adipose tissue inflammation.
Wang GX, Cho KW, Uhm M, Hu CR, Li S, Cozacov Z, Xu AE, Cheng JX, Saltiel AR, Lumeng CN, Lin JD. Wang GX, et al. Diabetes. 2014 Apr;63(4):1340-52. doi: 10.2337/db13-1139. Epub 2013 Dec 30. Diabetes. 2014. PMID: 24379350 Free PMC article. - SWI/SNF complex subunit BAF60a represses hepatic ureagenesis through a crosstalk between YB-1 and PGC-1α.
Zhang W, Dong Z, Xu M, Zhang S, Liu C, Chen S. Zhang W, et al. Mol Metab. 2020 Feb;32:85-96. doi: 10.1016/j.molmet.2019.12.007. Epub 2019 Dec 20. Mol Metab. 2020. PMID: 32029232 Free PMC article.
References
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. - PubMed
- Calfee-Mason KG, Spear BT, Glauert HP. Vitamin E inhibits hepatic NF-kappaB activation in rats administered the hepatic tumor promoter, phenobarbital. J Nutr. 2002;132:3178–3185. - PubMed
- Christian M, White R, Parker MG. Metabolic regulation by the nuclear receptor corepressor RIP140. Trends Endocrinol Metab. 2006;17:243–250. - PubMed
- Debril MB, Gelman L, Fayard E, Annicotte JS, Rocchi S, Auwerx J. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J Biol Chem. 2004;279:16677–16686. - PubMed
- Eaton S. Control of mitochondrial beta-oxidation flux. Prog Lipid Res. 2002;41:197–239. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK077086-02/DK/NIDDK NIH HHS/United States
- K01 DK065584-04/DK/NIDDK NIH HHS/United States
- K01 DK065584/DK/NIDDK NIH HHS/United States
- R01 DK077086-01A1/DK/NIDDK NIH HHS/United States
- DK077086/DK/NIDDK NIH HHS/United States
- DK065584/DK/NIDDK NIH HHS/United States
- R01 DK077086/DK/NIDDK NIH HHS/United States
- K01 DK065584-05/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases