PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks - PubMed (original) (raw)
Review
PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks
Lindsey A Baker et al. Mutat Res. 2008.
Abstract
Histone covalent modifications regulate many, if not all, DNA-templated processes, including gene expression and DNA damage response. The biological consequences of histone modifications are mediated partially by evolutionarily conserved "reader/effector" modules that bind to histone marks in a modification- and context-specific fashion and subsequently enact chromatin changes or recruit other proteins to do so. Recently, the Plant Homeodomain (PHD) finger has emerged as a class of specialized "reader" modules that, in some instances, recognize the methylation status of histone lysine residues, such as histone H3 lysine 4 (H3K4). While mutations in catalytic enzymes that mediate the addition or removal of histone modifications (i.e., "writers" and "erasers") are already known to be involved in various human diseases, mutations in the modification-specific "reader" proteins are only beginning to be recognized as contributing to human diseases. For instance, point mutations, deletions or chromosomal translocations that target PHD fingers encoded by many genes (such as recombination activating gene 2 (RAG2), Inhibitor of Growth (ING), nuclear receptor-binding SET domain-containing 1 (NSD1) and Alpha Thalassaemia and Mental Retardation Syndrome, X-linked (ATRX)) have been associated with a wide range of human pathologies including immunological disorders, cancers, and neurological diseases. In this review, we will discuss the structural features of PHD fingers as well as the diseases for which direct mutation or dysregulation of the PHD finger has been reported. We propose that misinterpretation of the epigenetic marks may serve as a general mechanism for human diseases of this category. Determining the regulatory roles of histone covalent modifications in the context of human disease will allow for a more thorough understanding of normal and pathological development, and may provide innovative therapeutic strategies wherein "chromatin readers" stand as potential drug targets.
Figures
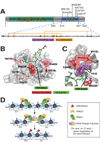
Figure 1. PHD fingers are “reader/effector” modules that recognize histone lysine methylation status
(A) A schematic model that illustrates the “writing”, “erasing” and “reading” of covalent modification marks on histone tails. (B–C) Two specialized subsets of PHD fingers specifically recognize and bind to the highly methylated-H3K4 (B) and unmodified H3K4 (C) respectively. The structures shown are PHD fingers of BPTF [11] and BHC80 [15], with PHD finger depicted in silver, H3K4 peptide in green and Zn2+ ion in cyan sphere. Side chains of the critical engaging residues are shown in purple. For simplicity, only modules engaging single marks are shown, and aspects involving multivalent engaging modules have been described elsewhere [7].
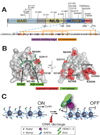
Figure 2. Missense mutations in the RAG2 PHD finger are associated with immunodeficiency syndromes
(A) Domain structure of RAG2 (NCBI accession number 187423896) showing disease-causing mutations found within the PHD finger. The amino acid sequence of the PHD finger is shown below with H3K4me-caging residues in purple and Zn2+ ion-coordinating residues in orange. (B) Co-crystal structure of the RAG2 PHD finger (in gray) and the H3K4me3 peptide (in green) [24]. Residues mutated in T-B-SCID or Omenn Syndrome are depicted in red, and Y415, which forms part of the aromatic channel, is in purple. (C) Closer view of mutations K440N and W416L as well as the RAG2 PHD aromatic channel with H3K4me3. (D) Schematic model for RAG2’s function in V(D)J recombination. Interactions between the RAG2 PHD and H3K4me3 help to recruit and/or stabilize RAG1/2 recombinases to appropriate V(D)J gene segments marked with this histone modification, where they create double-strand DNA breaks. Then DNA repair factors such as DNA-PKcs, Ku70/80, XRCC4 and DNA Ligase IV assists in ligation of the broken ends of two V(D)J gene segments, creating a functional gene segment. Note here the correlation between the disease-causing mutations and the “reading” function of the RAG2 PHD finger in engaging histone marks.
Figure 3. Mutations in the ING1b PHD finger isolated from cancers
(A) Domain structure of ING1b (p33ING1b, NCBI accession number 38201667) showing missense mutations reported in various solid cancers. The amino acid sequence of PHD finger is shown below, with the same color depiction as in Figure 2. SAID, SAP30-interacting domain [76]; NLS, nuclear localization signal; PBR, poly-basic region that mediates the binding to phosphoinositide [69]. (B) A structural model of f the ING1b PHD finger (in gray) associated with an H3K4me3 peptide (in green). Cancer-associated missense mutations as shown in red, and aromatic cage residues are in purple. This model is based on an co-crystal structure of the ING2 PHD finger and H3K4me3 peptide [13]. (C) A schematic model for ING-mediated cell cycle arrest. Upon DNA damage, the PHD finger of INGs that recognizes H3K4me3 marks enriched in promoters helps to stabilize the targeting of INGs to proliferative genes such as Cyclin wherein INGs subsequently recruit the SAP30/Sin3/HDAC complexes that remove acetyl marks from histone tails, repressing the transcription of proliferative genes and decelerating cell cycle progression.
Figure 4. Chromosomal aberrations that target PHD finger-containing proteins in hematopoietic malignancies
The NUP98 gene was found translocated to genes encoding the PHD finger-containing proteins JARID1A (panel A), PHF23 (panel B), NSD1 and NSD3 (panel C) in as subset of AML cases, and as a result, the leukemic fusion proteins harbor one or more PHD fingers. (D) MMSET, another PHD finger-containing protein, is commonly translocated in multiple myeloma. (E) Internal deletion (Δ) in MLL seen in T-cell leukaemia cases causes the disruption of the first PHD finger. Motif identities are specified in the boxed panel, and chromosomal translocation breakpoints are designated with arrows. The positions of nuclear localization signals (NLS) are only shown for panels A–B.
Similar articles
- Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.
Ryan R, Hill S. Ryan R, et al. Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article. - Enabling Systemic Identification and Functionality Profiling for Cdc42 Homeostatic Modulators.
Malasala S, Azimian F, Chen YH, Twiss JL, Boykin C, Akhtar SN, Lu Q. Malasala S, et al. bioRxiv [Preprint]. 2024 Jan 8:2024.01.05.574351. doi: 10.1101/2024.01.05.574351. bioRxiv. 2024. PMID: 38260445 Free PMC article. Updated. Preprint. - "It Is a Big Spider Web of Things": Sensory Experiences of Autistic Adults in Public Spaces.
MacLennan K, Woolley C, Andsensory E, Heasman B, Starns J, George B, Manning C. MacLennan K, et al. Autism Adulthood. 2023 Dec 1;5(4):411-422. doi: 10.1089/aut.2022.0024. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116051 Free PMC article. - Pharmacological treatments in panic disorder in adults: a network meta-analysis.
Guaiana G, Meader N, Barbui C, Davies SJ, Furukawa TA, Imai H, Dias S, Caldwell DM, Koesters M, Tajika A, Bighelli I, Pompoli A, Cipriani A, Dawson S, Robertson L. Guaiana G, et al. Cochrane Database Syst Rev. 2023 Nov 28;11(11):CD012729. doi: 10.1002/14651858.CD012729.pub3. Cochrane Database Syst Rev. 2023. PMID: 38014714 Free PMC article. Review.
Cited by
- Structural insights into acetylated histone ligand recognition by the BDP1 bromodomain of Plasmodium falciparum.
Singh AK, Phillips M, Alkrimi S, Tonelli M, Boyson SP, Malone KL, Nix JC, Glass KC. Singh AK, et al. Int J Biol Macromol. 2022 Dec 31;223(Pt A):316-326. doi: 10.1016/j.ijbiomac.2022.10.247. Epub 2022 Oct 31. Int J Biol Macromol. 2022. PMID: 36328269 Free PMC article. - Phf14, a novel regulator of mesenchyme growth via platelet-derived growth factor (PDGF) receptor-α.
Kitagawa M, Takebe A, Ono Y, Imai T, Nakao K, Nishikawa S, Era T. Kitagawa M, et al. J Biol Chem. 2012 Aug 10;287(33):27983-96. doi: 10.1074/jbc.M112.350074. Epub 2012 Jun 23. J Biol Chem. 2012. PMID: 22730381 Free PMC article. - The Structural Determinants behind the Epigenetic Role of Histone Variants.
Cheema MS, Ausió J. Cheema MS, et al. Genes (Basel). 2015 Jul 23;6(3):685-713. doi: 10.3390/genes6030685. Genes (Basel). 2015. PMID: 26213973 Free PMC article. Review. - High-throughput strategy to identify inhibitors of histone-binding domains.
Wagner EK, Albaugh BN, Denu JM. Wagner EK, et al. Methods Enzymol. 2012;512:161-85. doi: 10.1016/B978-0-12-391940-3.00008-1. Methods Enzymol. 2012. PMID: 22910207 Free PMC article. - Whole Exome Sequencing Identifies PHF14 Mutations in Neurocytoma and Predicts Responsivity to the PDGFR Inhibitor Sunitinib.
Zhang D, Yong W, Movassaghi M, Rodriguez FJ, Yang I, McKeever P, Qian J, Li JY, Mao Q, Newell KL, Green RM, Welsh CT, Heaney AP. Zhang D, et al. Biomedicines. 2022 Nov 8;10(11):2842. doi: 10.3390/biomedicines10112842. Biomedicines. 2022. PMID: 36359362 Free PMC article.
References
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. - PubMed
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. - PubMed
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 GM053512-30/GM/NIGMS NIH HHS/United States
- GM 53512/GM/NIGMS NIH HHS/United States
- T32 CA009673/CA/NCI NIH HHS/United States
- CA09673/CA/NCI NIH HHS/United States
- R37 GM053512/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases