Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data - PubMed (original) (raw)
Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data
Raja Jothi et al. Nucleic Acids Res. 2008 Sep.
Abstract
ChIP-Seq, which combines chromatin immunoprecipitation (ChIP) with ultra high-throughput massively parallel sequencing, is increasingly being used for mapping protein-DNA interactions in-vivo on a genome scale. Typically, short sequence reads from ChIP-Seq are mapped to a reference genome for further analysis. Although genomic regions enriched with mapped reads could be inferred as approximate binding regions, short read lengths (approximately 25-50 nt) pose challenges for determining the exact binding sites within these regions. Here, we present SISSRs (Site Identification from Short Sequence Reads), a novel algorithm for precise identification of binding sites from short reads generated from ChIP-Seq experiments. The sensitivity and specificity of SISSRs are demonstrated by applying it on ChIP-Seq data for three widely studied and well-characterized human transcription factors: CTCF (CCCTC-binding factor), NRSF (neuron-restrictive silencer factor) and STAT1 (signal transducer and activator of transcription protein 1). We identified 26 814, 5813 and 73 956 binding sites for CTCF, NRSF and STAT1 proteins, respectively, which is 32, 299 and 78% more than that inferred previously for the respective proteins. Motif analysis revealed that an overwhelming majority of the identified binding sites contained the previously established consensus binding sequence for the respective proteins, thus attesting for SISSRs' accuracy. SISSRs' sensitivity and precision facilitated further analyses of ChIP-Seq data revealing interesting insights, which we believe will serve as guidance for designing ChIP-Seq experiments to map in vivo protein-DNA interactions. We also show that tag densities at the binding sites are a good indicator of protein-DNA binding affinity, which could be used to distinguish and characterize strong and weak binding sites. Using tag density as an indicator of DNA-binding affinity, we have identified core residues within the NRSF and CTCF binding sites that are critical for a stronger DNA binding.
Figures
Figure 1.
Schematic overview of SISSRs algorithm. (A) Sequenced short reads (typically ∼25–50 bp) from ChIP-Seq experiments are first mapped onto the reference genome. The mapped reads are then used to estimate statistical parameters, which include the estimation of the average length F of sequenced DNA fragments. (B) The entire reference genome along with mapped reads is scanned using overlapping windows of size w base pairs (overlapping not shown in the figure for clarity), and the net tag count (ci) for every window i is calculated. Every transition point (t) is a candidate binding site, and needs to satisfy a set of estimated as well as user-set thresholds in order to be classified as a true binding site.
Figure 2.
Distribution of binding sites across the genome. RefSeq genes were used as reference. The 5 kb region upstream of transcription start site was defined as the promoter.
Figure 3.
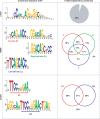
Accuracy of inferred binding sites. Consensus binding motifs for CTCF, NRSF and STAT1 binding sites are shown in the left panel. The right panel shows the percentage of identified binding sites containing the consensus binding motif(s).
Figure 4.
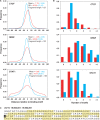
Resolution of inferred binding sites. (A) Distribution of the distance between SISSRs-inferred binding site and the middle of the nearest canonical motif occurrence. The red curve is for all binding sites, and the blue curve is for high-scoring binding sites. About 75% of all CTCF, NRSF and STAT1 sites were within 18, 27 and 51 bp of the nearest consensus motif, respectively (90% within 31, 52 and 73 bp, respectively). The resolution is much higher for high-scoring binding sites (90% within 20, 19 and 45 bp, respectively). (B) Distribution of the number of canonical motifs within the 200-bp region centered on identified binding sites. The coloring scheme is the same as that used in A. (C) A 200-bp region centered on a CTCF binding site (highlighted in black) with high tag density. Nine instances of core CTCF motif (10−8 < P < 0.0025) are highlighted in yellow.
Figure 5.
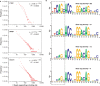
Tag density at a binding site is an indicator of binding affinity. (A) Distribution of the number of tags within the 200-bp region centered on binding sites exhibits a power-law behavior. (B) Motif analysis on four mutually exclusive sets of 500 NRSF binding sites each reveals a positive correlation between the information content of first 6 nt of the NRSF consensus motif and the average tag density. See
Supplementary Figure 2
for an analysis of CTCF binding sites.
Figure 6.
Overcoming ChIP-Seq limitations. (A) Nonspecific reads due to antibody non-specificity and/or sequencing errors could be controlled for by adjusting the window size w. (B) SISSRs provides an option to identify binding sites with tags mapped to only the sense or antisense strand, which otherwise may not have been identified (site A). Steps are taken to make sure that tags corresponding to site B are not accounted while identifying site A: the right most sense tag i for site A should be at least a fragment length F away from the nearest sense tag (j) and at least 2_F_ away from the nearest antisense tag k. (C) False positives due to indirect DNA binding via protein–protein interaction is a cause for concern, and are difficult to avoid as it is very difficult to determine whether the binding to the DNA is direct or indirect.
Similar articles
- De novo motif identification improves the accuracy of predicting transcription factor binding sites in ChIP-Seq data analysis.
Boeva V, Surdez D, Guillon N, Tirode F, Fejes AP, Delattre O, Barillot E. Boeva V, et al. Nucleic Acids Res. 2010 Jun;38(11):e126. doi: 10.1093/nar/gkq217. Epub 2010 Apr 7. Nucleic Acids Res. 2010. PMID: 20375099 Free PMC article. - Discovering transcription factor binding sites in highly repetitive regions of genomes with multi-read analysis of ChIP-Seq data.
Chung D, Kuan PF, Li B, Sanalkumar R, Liang K, Bresnick EH, Dewey C, Keleş S. Chung D, et al. PLoS Comput Biol. 2011 Jul;7(7):e1002111. doi: 10.1371/journal.pcbi.1002111. Epub 2011 Jul 14. PLoS Comput Biol. 2011. PMID: 21779159 Free PMC article. - An integrated pipeline for the genome-wide analysis of transcription factor binding sites from ChIP-Seq.
Mercier E, Droit A, Li L, Robertson G, Zhang X, Gottardo R. Mercier E, et al. PLoS One. 2011 Feb 16;6(2):e16432. doi: 10.1371/journal.pone.0016432. PLoS One. 2011. PMID: 21358819 Free PMC article. - Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond.
Mundade R, Ozer HG, Wei H, Prabhu L, Lu T. Mundade R, et al. Cell Cycle. 2014;13(18):2847-52. doi: 10.4161/15384101.2014.949201. Cell Cycle. 2014. PMID: 25486472 Free PMC article. Review. - Genome Wide Approaches to Identify Protein-DNA Interactions.
Ma T, Ye Z, Wang L. Ma T, et al. Curr Med Chem. 2019;26(42):7641-7654. doi: 10.2174/0929867325666180530115711. Curr Med Chem. 2019. PMID: 29848263 Review.
Cited by
- Kinetics and functional diversity among the five members of the NADP-malic enzyme family from Zea mays, a C4 species.
Alvarez CE, Saigo M, Margarit E, Andreo CS, Drincovich MF. Alvarez CE, et al. Photosynth Res. 2013 May;115(1):65-80. doi: 10.1007/s11120-013-9839-9. Epub 2013 May 7. Photosynth Res. 2013. PMID: 23649167 - An Overview of Genome Organization and How We Got There: from FISH to Hi-C.
Fraser J, Williamson I, Bickmore WA, Dostie J. Fraser J, et al. Microbiol Mol Biol Rev. 2015 Sep;79(3):347-72. doi: 10.1128/MMBR.00006-15. Microbiol Mol Biol Rev. 2015. PMID: 26223848 Free PMC article. Review. - Harnessing genetic engineering to drive economic bioproduct production in algae.
Gupta A, Kang K, Pathania R, Saxton L, Saucedo B, Malik A, Torres-Tiji Y, Diaz CJ, Dutra Molino JV, Mayfield SP. Gupta A, et al. Front Bioeng Biotechnol. 2024 Jan 29;12:1350722. doi: 10.3389/fbioe.2024.1350722. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 38347913 Free PMC article. Review. - DeepD2V: A Novel Deep Learning-Based Framework for Predicting Transcription Factor Binding Sites from Combined DNA Sequence.
Deng L, Wu H, Liu X, Liu H. Deng L, et al. Int J Mol Sci. 2021 May 24;22(11):5521. doi: 10.3390/ijms22115521. Int J Mol Sci. 2021. PMID: 34073774 Free PMC article. - Transcriptomics in type 2 diabetes: Bridging the gap between genotype and phenotype.
Jenkinson CP, Göring HH, Arya R, Blangero J, Duggirala R, DeFronzo RA. Jenkinson CP, et al. Genom Data. 2015 Dec 17;8:25-36. doi: 10.1016/j.gdata.2015.12.001. eCollection 2016 Jun. Genom Data. 2015. PMID: 27114903 Free PMC article.
References
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. - PubMed
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. - PubMed
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. - PubMed
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous