Tyr130 phosphorylation triggers Syk release from antigen receptor by long-distance conformational uncoupling - PubMed (original) (raw)
Tyr130 phosphorylation triggers Syk release from antigen receptor by long-distance conformational uncoupling
Yajie Zhang et al. Proc Natl Acad Sci U S A. 2008.
Abstract
The Syk protein-tyrosine kinase plays a major role in signaling through the B cell receptor for antigen (BCR). Syk binds the receptor via its tandem pair of SH2 domains interacting with a doubly phosphorylated immunoreceptor tyrosine-based activation motif (dp-ITAM) of the BCR complex. Upon phosphorylation of Tyr-130, which lies between the two SH2 domains distant to the phosphotyrosine binding sites, Syk dissociates from the receptor. To understand the structural basis for this dissociation, we investigated the structural and dynamic characteristics of the wild type tandem SH2 region (tSH2) and a variant tandem SH2 region (tSH2(pm)) with Tyr-130 substituted by Glu to permanently introduce a negative charge at this position. NMR heteronuclear relaxation experiments, residual dipolar coupling measurements and analytical ultracentrifugation revealed substantial differences in the hydrodynamic behavior of tSH2 and tSH2(pm). Although the two SH2 domains in tSH2 are tightly associated, the two domains in tSH2(pm) are partly uncoupled and tumble in solution with a faster correlation time. In addition, the equilibrium dissociation constant for the binding of tSH2(pm) to dp-ITAM (1.8 microM) is significantly higher than that for the interaction between dp-ITAM and tSH2 but is close to that for a singly tyrosine-phosphorylated peptide binding to a single SH2 domain. Experimental data and hydrodynamic calculations both suggest a loss of domain-domain contacts and change in relative orientation upon the introduction of a negative charge on residue 130. A long-distance structural mechanism by which the phosphorylation of Y130 negatively regulates the interaction of Syk with immune receptors is proposed.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
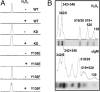
The binding of Syk to a phosphorylated ITAM is inhibited by the autophosphorylation of Y130. (A) Syk-deficient DT40 cells stably expressing Myc-tagged murine Syk (WT), Syk(K396R) (KD), Syk(Y130E) or Syk(Y130F) were treated without (−) or with (+) the phosphatase inhibitor H2O2 (10 mM). Detergent lysates were adsorbed to an immobilized phosphopeptide corresponding to the doubly phosphorylated CD79a ITAM. Bound Syk, plotted here, was detected by Western blot analysis with an anti-Syk antibody and quantified by densitometry. (B) Myc-tagged Syk was immunoprecipitated with anti-Myc antibodies from lysates of cells preincubated in [32P]orthophosphate and treated with H2O2 (Upper) or anti-IgM antibodies (Lower). The recovered protein was digested with trypsin and the resulting phosphopeptides separated electrophoretically and detected by autoradiography (Upper) or with a phosphoimager (Lower). The region of the gel containing the smaller and more rapidly migrating phosphopeptides is illustrated. The positions of the phosphotyrosines within each peptide are numbered based on the murine Syk sequence.
Fig. 2.
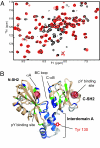
Spectral differences between tSH2 and tSH2pm. (A) Overlay of part of the 1H-15N HSQC spectra of tSH2 (red) and tSH2pm (black) recorded at 600 MHz, 20°C. A number of peaks from residues in interdomain A and at the SH2 domain interface are observed in the tSH2pm spectrum, but missing in the tSH2 spectrum. (B) Spectral differences are mapped onto the crystal structure for the Syk tSH2 complexed with the phosphorylated CD3ε-ITAM (PDB 1A81). Residues are colored based on HSQC peaks: observed for tSH2 but not tSH2pm (blue), a difference in chemical shift >0.04 ppm (green), or <0.04 ppm (tan), not observed (gray). Y130 is shown in sticks. Spheres show the location of the two phosphotyrosine binding pockets. The amide group chemical shift difference between the two protein constructs, Δ1H+15N, is calculated from the frequency difference, δi, of nucleus i (33): Δ1H+15N= .
.
Fig. 3.
15N relaxation data (600 MHz, 293 K) for tSH2 (black) and tSH2pm (gray). Heteronuclear NOE values (Lower) and _R_2/_R_1 values (Upper) are plotted on individual residues basis. The secondary structure elements in the Lower are as follows. β-strand (Black): βA (residues 15–17, 168–171), βB (residues 38–43, 191–196), βC (residues 51–57, 202–209), βDβD′ (residues 60–69, 212–221), βE (residues 74–76, 225–228), βF (residues 80–82, 232–234), βG (residues 105–107, 257–259). α-helix (Gray): αA (residues 22–31, 175–183) αB (residues 85–93,237–246). Errors were calculated by pairwise root mean square deviation of duplicate experiments.
Fig. 4.
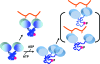
Model of a long-distance mechanism for Y130 (red sphere) phosphorylation to down-regulate association of Syk with the antigen receptor. Phosphorylation destabilizes interdomain A structure, which alters SH2-SH2 domain orientation and retains an increased distance between pY binding sites beyond the limit permitted for bifunctional binding of ITAM.
Similar articles
- Entropic allostery dominates the phosphorylation-dependent regulation of Syk tyrosine kinase release from immunoreceptor tyrosine-based activation motifs.
Feng C, Roy A, Post CB. Feng C, et al. Protein Sci. 2018 Oct;27(10):1780-1796. doi: 10.1002/pro.3489. Epub 2018 Oct 2. Protein Sci. 2018. PMID: 30051939 Free PMC article. - Crystal structure and NMR studies of the apo SH2 domains of ZAP-70: two bikes rather than a tandem.
Folmer RH, Geschwindner S, Xue Y. Folmer RH, et al. Biochemistry. 2002 Dec 3;41(48):14176-84. doi: 10.1021/bi026465e. Biochemistry. 2002. PMID: 12450381 - The tyrosine activation motif as a target of protein tyrosine kinases and SH2 domains.
Flaswinkel H, Barner M, Reth M. Flaswinkel H, et al. Semin Immunol. 1995 Feb;7(1):21-7. doi: 10.1016/1044-5323(95)90004-7. Semin Immunol. 1995. PMID: 7612891 Review. - Syk and pTyr'd: Signaling through the B cell antigen receptor.
Geahlen RL. Geahlen RL. Biochim Biophys Acta. 2009 Jul;1793(7):1115-27. doi: 10.1016/j.bbamcr.2009.03.004. Epub 2009 Mar 21. Biochim Biophys Acta. 2009. PMID: 19306898 Free PMC article. Review.
Cited by
- The SYK tyrosine kinase: a crucial player in diverse biological functions.
Mócsai A, Ruland J, Tybulewicz VL. Mócsai A, et al. Nat Rev Immunol. 2010 Jun;10(6):387-402. doi: 10.1038/nri2765. Nat Rev Immunol. 2010. PMID: 20467426 Free PMC article. Review. - Molecular mechanism of selective recruitment of Syk kinases by the membrane antigen-receptor complex.
Bond PJ, Faraldo-Gómez JD. Bond PJ, et al. J Biol Chem. 2011 Jul 22;286(29):25872-81. doi: 10.1074/jbc.M111.223321. Epub 2011 May 21. J Biol Chem. 2011. PMID: 21602568 Free PMC article. - TULA-2 Protein Phosphatase Suppresses Activation of Syk through the GPVI Platelet Receptor for Collagen by Dephosphorylating Tyr(P)346, a Regulatory Site of Syk.
Reppschläger K, Gosselin J, Dangelmaier CA, Thomas DH, Carpino N, McKenzie SE, Kunapuli SP, Tsygankov AY. Reppschläger K, et al. J Biol Chem. 2016 Oct 21;291(43):22427-22441. doi: 10.1074/jbc.M116.743732. Epub 2016 Sep 8. J Biol Chem. 2016. PMID: 27609517 Free PMC article. - Insights into the allosteric regulation of Syk association with receptor ITAM, a multi-state equilibrium.
Feng C, Post CB. Feng C, et al. Phys Chem Chem Phys. 2016 Feb 17;18(8):5807-18. doi: 10.1039/c5cp05417f. Phys Chem Chem Phys. 2016. PMID: 26468009 Free PMC article. - A reevaluation of the spleen tyrosine kinase (SYK) activation mechanism.
Mansueto MS, Reens A, Rakhilina L, Chi A, Pan BS, Miller JR. Mansueto MS, et al. J Biol Chem. 2019 May 10;294(19):7658-7668. doi: 10.1074/jbc.RA119.008045. Epub 2019 Mar 28. J Biol Chem. 2019. PMID: 30923129 Free PMC article.
References
- Turner M, Schweighoffer E, Colucci F, Di Santo JP, Tybulewicz VL. Tyrosine kinase SYK: Essential functions for immunoreceptor signalling. Immunol Today. 2000;21:148–154. - PubMed
- Reth M. Antigen receptor tail clue. Nature. 1989;338:383–384. - PubMed
- Grucza RA, Futterer K, Chan AC, Waksman G. Thermodynamic study of the binding of the tandem-SH2 domain of the Syk kinase to a dually phosphorylated ITAM peptide: Evidence for two conformers. Biochemistry. 1999;38:5024–5033. - PubMed
- Ottinger EA, Botfield MC, Shoelson SE. Tandem SH2 domains confer high specificity in tyrosine kinase signaling. J Biol Chem. 1998;273:729–735. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous