Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma - PubMed (original) (raw)
. 2008 Aug 12;14(2):123-34.
doi: 10.1016/j.ccr.2008.07.005.
Vivi M Heine, Junhao Mao, Alvin T Kho, Allison K Dillon, Young-Goo Han, Emmanuelle Huillard, Tao Sun, Azra H Ligon, Ying Qian, Qiufu Ma, Arturo Alvarez-Buylla, Andrew P McMahon, David H Rowitch, Keith L Ligon
Affiliations
- PMID: 18691547
- PMCID: PMC2597270
- DOI: 10.1016/j.ccr.2008.07.005
Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma
Ulrich Schüller et al. Cancer Cell. 2008.
Abstract
Whether the brain tumor medulloblastoma originates from stem cells or restricted progenitor cells is unclear. To investigate this, we activated oncogenic Hedgehog (Hh) signaling in multipotent and lineage-restricted central nervous system (CNS) progenitors. We observed that normal unipotent cerebellar granule neuron precursors (CGNPs) derive from hGFAP(+) and Olig2(+) rhombic lip progenitors. Hh activation in a spectrum of early- and late-stage CNS progenitors generated similar medulloblastomas, but not other brain cancers, indicating that acquisition of CGNP identity is essential for tumorigenesis. We show in human and mouse medulloblastoma that cells expressing the glia-associated markers Gfap and Olig2 are neoplastic and retain features of embryonic-type granule lineage progenitors. Thus, oncogenic Hh signaling promotes medulloblastoma from lineage-restricted granule cell progenitors.
Figures
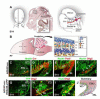
Figure 1. Identification of an Olig2+ progenitor population in the rostral RL
(A) The cerebellar anlagen at E14. The VZ and RL serve as mitotic niches at this age. The EGL is formed by proliferating CGNP that have left the RL. (B) At P7, the developing CB is divided into 10 lobes with defined cortical layers. (C-E) Sagittal sections at E14 in hGFAP-cre mice showing co-localization of Nestin, cre and Pax6 proteins in progenitor cells of the VZ and rRL. (F) Olig2 expression is largely restricted to dorsal rRL. (G, H) Fate mapping experiments (hGFAP-cre × ROSA26-eYFPFl/Fl mice) reveals YFP in the EGL and a subset of RL cells that express Olig2 (arrows). (I) Olig2+ RL precursors co-label with the CGNP marker Pax6; some single Pax6+ precursors in the VZ were observed (arrows). (J) Summary of cells in the RL region at E14 showing relative positions of radial glia (yellow/green), Pax6(blue) +, Olig2(red)+ and Pax6+Olig2(purple)+ cells. BG, Bergmann glia; CP, choroid plexus; GNP, granule neuron precursors; ML, molecular layer; PL, Purkinje cell layer.
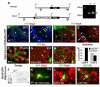
Figure 2. A subset of cerebellar granule neurons derive from Olig2+ progenitors
(A)Olig2-tva-cre allele and genotyping of the respective mutant mice using PCR to generate wild type 429bp and mutant (cre) 446bp products. (B-I) Fate mapping of Olig2-expressing cells with ROSA26-eYFPFl/Fl reporter. Co-localization of YFP and Olig2 proteins in sagittal sections of P7 cerebella reveals that YFP is expressed in oligodendrocytes within the cerebellar WM (yellow arrows in C) as well as in occasional cells within the EGL (white arrows). YFP-Pax6 labeling confirms the granule lineage character of YFP-expressing EGL cells (D, E, arrows). The IGL of P21 cerebella demonstrates lobe-specific differences in Olig2 fate mapping. (G) The IGL of Lobe II contains YFP+ cells that predominantly co-express Olig2 (arrow and cell shown, inset) but not the GC marker Zic. In contrast, the IGL of Lobe X (H) contains numerous YFP/Zic double-positive cells (arrow and inset). (I) Mean percentage (+/-SEM) of Olig2+YFP+ and Zic1+YFP+ double-positive cells in the IGL of lobe II and X. (J-M) In utero fate mapping of Olig2-expressing cells by RCAS-GFP virus injection into the rRL of E15.5 Olig2-tva-cre mice. Analysis of the CB at P7 reveals GFP to be expressed in Olig2+ (K) WM cells, as well as Pax6+ (L) and Zic+ (M) cells of the IGL in Lobe X.
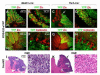
Figure 3. Development of medulloblastoma but not glioma from hGFAP+ and Olig2+ precursors
Targeting hGFAP+ and Olig2+ precursor cells with SmoM2 results in MB (A-H). While neoplastic transformation of hGFAP+ cells affects the entire cerebellar cortex (A-D), MBs from _Olig2_-expressing precursors are restricted to posterior cerebellar regions (E-H). Although _hGFAP_-cre and _Olig2_-cre are broadly expressed in all parts of the CNS, neoplastic lesions due to SmoM2 are restricted to the CB. Forebrain sections including the subventricular zone (SVZ) in adult animals appear normal (I-L). Transplantation of Olig2-tva-cre:SmoM2 tumor cells into SCID mice gives rise tumors both in hindbrain (M,N) and forebrain (O,P) regions. A, E and I-P are frontal, B-D and F-H are sagittal sections stained with H&E. C and D are high power magnifications from B, so are G and H from F. Asterisks mark lateral ventricles in I, K and O and the IVth ventricle in M. BS, brainstem.
Figure 4. Late stage unipotent CGNP are competent to produce medulloblastoma
(A-H) Cre expression driven by Math1 or Tlx3 promoter sequences is normally restricted to granule lineage cell as shown by crosses with Rosa26-eYFP conditional reporter mice. (C, D, G) GC fate-mapped by _Tlx3_-cre are restricted to Lobes VI-IX. (F, H) In contrast, Calbindin-expressing PN do not derive from Math1+ or Tlx3+ precursors. (I-L) Both Math1+ and Tlx3+ granule neurons give rise to MB after activation of SmoM2, and Tlx3-cre driven tumors obeyed posterior restriction consistent with fate mapping (compare arrows in C, K).
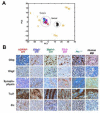
Figure 5. Medulloblastoma generated from diverse progenitor cell origins leads to a convergent phenotypic endpoint
(A) Principal component analysis of global gene expression in conditional MB models [Math1(green)-, Tlx3(magenta)-, hGFAP(red)-, Olig2(blue)-cre:SmoM2 and Ptc+/- (turquoise)] and samples from normal littermate control CB (black circles) are mapped onto normal cerebellar developmental space as defined by a 7000 member, rank-normalized gene set (Kho et al., 2004), from developing CB at stages P1, 3, 5, 7, 10, 15, 21, 30, 50 and 60 (orange numbers). Tumors were related to early rather than late developmental stages of the CB as we have reported for human MB (Kho et al., 2004). (B) Immunohistochemical analysis confirms all murine tumors exhibit a similar immunohistochemical staining pattern to each other as well as human MB with respect to standard neuronal and glial neuropathological markers. Results are representative of at least two tumor samples.
Figure 6. Olig2- and Gfap-expressing cells in medulloblastoma are neoplastic and have features in common with immature granule lineage precursors
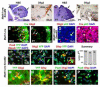
(A, C) In Math1-cre:SmoM2 animals, morphologic evidence hyperplasia is predominantly observed in the EGL, but not RL or VZ, at P0 and is more apparent by P7. (B, D) Greatly increased numbers of Olig2+ cells were observed in hyperplastic regions of the EGL--but not RL--of P0 and P7 Math1-cre:SmoM2 mice. (E, F) Expression of cre proteins in Math1-cre mice co-localized with Olig2 at E18 but not P7. (G) Olig2 co-expression was detected in only ∼2% of pH3+ cells in tumors. (H) In contrast, ∼98% of pH3+ cells co-labeled with cre proteins. (I-K) Phenotype of acutely disassociated cells and (insets) cells cultured from MB for four weeks. While most of dissociated Math1-cre:SmoM2 tumor cells express the GC markers Pax6 or (inset) NeuN (I), some express Olig2 (J) and Gfap (K). SmoM2 is fused to YFP, which therefore marks tumor cells. (L) Cell counting reveals that more than 90% of cells expressing Gfap, Olig2 and Pax6 are tumor cells, as shown by co-expression of YFP. (M, N) Immunohistochemistry of tumor sections demonstrates similar results. (O) 81% of Olig2+ and (P) 100% of Gfap+ cells in tumors expressed the CGNP marker Pax6.
Figure 7. OLIG2 is expressed in human medulloblastoma
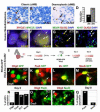
(A-D) OLIG2 expression in human classic and desmoplastic MB and percentage of tumors of each class that contain OLIG+ cells (also see Suppl. Tab 1). (E, F) Combined FISH and immunohistochemical analysis reveals the presence of tumor specific C-MYC genomic aberrations (Herms et al., 2000) within OLIG2+ cells (yellow nuclei) of human MB (_n_=2). C-MYC aberrations were identical in OLIG2+ and OLIG2- tumor cells. Note combined copy number gain (red signals, 5′ C-MYC) and structural aberration (loss of green signals, 3′ C-MYC), as detected using break apart probe set. Normal MYC loci show merged colors as (focal yellow signals). (G, H) OLIG2 co-localizes with NEUN, and the proliferation marker Ki67. (I) Scheme for experiments using RCAS-GFP infection of Olig2-tva-cre:SmoM2 tumor cells. (J-M) Double-fluorescence images show expression of GFP that co-localizes with immunoreactivity using antibodies against Olig2 and GC lineage markers. Note that while Olig2-tva-cre:SmoM2 express YFP the signal is very weak/undetectable compared with GFP encoded by RCAS virus, as shown (J-M). (N) Summary of findings at Day 6 in GFP-labeled cells. (O-Q) Analysis of acutely disassociated Olig2-tva-cre:SmoM2 tumor (Day 0). Of Olig2+ cells, 66% co-labeled with Pax6 and 12% with NeuN, such cells represented 4% and 1% of total cells in the tumor, respectively.
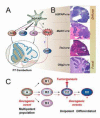
Figure 8. Tumor competent cells of the CGNP lineage can give rise to diffuse and focal forms of medulloblastoma
(A) Scheme showing P7 neuronal progeny deriving from _hGFAP_- and _Olig2_-expressing progenitors of the embryonic CB. hGFAP+ cells produce the majority of granule cell precursors (GNP) as well as astrocytes (Glia) and IN. While some Olig2+ cells derive from hGFAP+ cells (black dashed arrow), additional sources are possible. Olig2+ cells of the rRL produce a subset of GNP in cerebellar lobes IX and X, and also give rise to IN and oligodendrocytes (Glia) in all lobes (red arrows). The blue dashed arrow indicates the possibility of GNP de-differentiation in the context of Hh-induced MB. (B) Activation of SmoM2 in GNP using _hGFAP_- or Math1-cre produced diffuse MB involving most cerebellar lobes and the vermis, while focal, posterior-lateral MB was produced from the Olig2-cre driver; Tlx3-cre produced tumors with posterior restriction. (C) Possible cellular pathways of tumorigenesis. Introduction of an oncogene (red arrowheads) into multipotent (A, giving rise to distinct progeny types B1, B2, B3), unipotent (C2) or even differentiated (D2) subtypes of a tumor-competent lineage (e.g., GNP of CB) might result in a uniform tumor phenotype providing only one stage (C2) is capable of rapid expansion in response to the signal. This seems the simplest model to fit our collective findings; however, tumor propagation from an A or B2 cell cannot be ruled out. The later possibility of de-differentiation from D2 > C2 is speculative and not supported by the present work.
Comment in
- Even cancers want commitment: lineage identity and medulloblastoma formation.
Eberhart CG. Eberhart CG. Cancer Cell. 2008 Aug 12;14(2):105-7. doi: 10.1016/j.ccr.2008.07.011. Cancer Cell. 2008. PMID: 18691544 Free PMC article.
Similar articles
- Sonic hedgehog-associated medulloblastoma arising from the cochlear nuclei of the brainstem.
Grammel D, Warmuth-Metz M, von Bueren AO, Kool M, Pietsch T, Kretzschmar HA, Rowitch DH, Rutkowski S, Pfister SM, Schüller U. Grammel D, et al. Acta Neuropathol. 2012 Apr;123(4):601-14. doi: 10.1007/s00401-012-0961-0. Epub 2012 Feb 21. Acta Neuropathol. 2012. PMID: 22349907 - Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells.
Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, Schüller U, Machold R, Fishell G, Rowitch DH, Wainwright BJ, Wechsler-Reya RJ. Yang ZJ, et al. Cancer Cell. 2008 Aug 12;14(2):135-45. doi: 10.1016/j.ccr.2008.07.003. Cancer Cell. 2008. PMID: 18691548 Free PMC article. - Even cancers want commitment: lineage identity and medulloblastoma formation.
Eberhart CG. Eberhart CG. Cancer Cell. 2008 Aug 12;14(2):105-7. doi: 10.1016/j.ccr.2008.07.011. Cancer Cell. 2008. PMID: 18691544 Free PMC article. - RBP-J is not required for granule neuron progenitor development and medulloblastoma initiated by Hedgehog pathway activation in the external germinal layer.
Julian E, Hallahan AR, Wainwright BJ. Julian E, et al. Neural Dev. 2010 Oct 15;5:27. doi: 10.1186/1749-8104-5-27. Neural Dev. 2010. PMID: 20950430 Free PMC article. - Olig2 transcription factor in the developing and injured forebrain; cell lineage and glial development.
Ono K, Takebayashi H, Ikenaka K. Ono K, et al. Mol Cells. 2009 Apr 30;27(4):397-401. doi: 10.1007/s10059-009-0067-2. Epub 2009 Apr 13. Mol Cells. 2009. PMID: 19390819 Review.
Cited by
- Sonic hedgehog signalling pathway in CNS tumours: its role and therapeutic implications.
Wireko AA, Ben-Jaafar A, Kong JSH, Mannan KM, Sanker V, Rosenke SL, Boye ANA, Nkrumah-Boateng PA, Poornaselvan J, Shah MH, Abdul-Rahman T, Atallah O. Wireko AA, et al. Mol Brain. 2024 Nov 20;17(1):83. doi: 10.1186/s13041-024-01155-w. Mol Brain. 2024. PMID: 39568072 Free PMC article. Review. - Modulation of Stemness and Differentiation Regulators by Valproic Acid in Medulloblastoma.
Freire NH, Herlinger AL, Vanini J, Dalmolin M, Fernandes MAC, Nör C, Ramaswamy V, de Farias CB, Brunetto AT, Brunetto AL, Gregianin LJ, da Cunha Jaeger M, Taylor MD, Roesler R. Freire NH, et al. bioRxiv [Preprint]. 2024 Sep 25:2024.09.23.614476. doi: 10.1101/2024.09.23.614476. bioRxiv. 2024. PMID: 39386542 Free PMC article. Preprint. - Understanding the autophagic functions in cancer stem cell maintenance and therapy resistance.
Niharika, Garg M. Niharika, et al. Expert Rev Mol Med. 2024 Oct 8;26:e23. doi: 10.1017/erm.2024.23. Expert Rev Mol Med. 2024. PMID: 39375840 Free PMC article. Review. - Thyroid hormone suppresses medulloblastoma progression through promoting terminal differentiation of tumor cells.
Yang Y, Valdés-Rives SA, Liu Q, Gao T, Burudpakdee C, Li Y, Tan J, Tan Y, Koch CA, Rong Y, Houser SR, Wei S, Cai KQ, Wu J, Cheng SY, Wechsler-Reya R, Yang ZJ. Yang Y, et al. Cancer Cell. 2024 Aug 12;42(8):1434-1449.e5. doi: 10.1016/j.ccell.2024.07.008. Cancer Cell. 2024. PMID: 39137728 - Permanent deconstruction of intracellular primary cilia in differentiating granule cell neurons.
Ott CM, Constable S, Nguyen TM, White K, Lee WA, Lippincott-Schwartz J, Mukhopadhyay S. Ott CM, et al. J Cell Biol. 2024 Oct 7;223(10):e202404038. doi: 10.1083/jcb.202404038. Epub 2024 Aug 13. J Cell Biol. 2024. PMID: 39137043 Free PMC article.
References
- Abraham H, Tornoczky T, Kosztolanyi G, Seress L. Cell formation in the cortical layers of the developing human cerebellum. Int. J. Dev. Neurosci. 2001;19:53–62. - PubMed
- Ahn S, Joyner AL. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005;437:894–897. - PubMed
- Altmann J, Bayer SA. Development of the Cerebellar System in Relation to its Evolution, Structure and Functions. CRC Press; New York: 1997.
- Barabe F, Kennedy JA, Hope KJ, Dick JE. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316:600–604. - PubMed
- Ben Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NS040828/NS/NINDS NIH HHS/United States
- R01 NS033642/NS/NINDS NIH HHS/United States
- P01 NS040828/NS/NINDS NIH HHS/United States
- NS047527/NS/NINDS NIH HHS/United States
- R37 NS033642/NS/NINDS NIH HHS/United States
- P50 NS040828/NS/NINDS NIH HHS/United States
- K08 NS047213-01A1/NS/NINDS NIH HHS/United States
- R01 NS047527-04/NS/NINDS NIH HHS/United States
- R01 NS047527-01A1/NS/NINDS NIH HHS/United States
- NS033642/NS/NINDS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- K08 NS047213/NS/NINDS NIH HHS/United States
- R01 NS047527/NS/NINDS NIH HHS/United States
- R01 NS047527-03/NS/NINDS NIH HHS/United States
- R01 NS047527-02/NS/NINDS NIH HHS/United States
- NS047213/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous