A protein domain-based interactome network for C. elegans early embryogenesis - PubMed (original) (raw)
. 2008 Aug 8;134(3):534-45.
doi: 10.1016/j.cell.2008.07.009.
Zoltan Maliga, Niels Klitgord, Na Li, Irma Lemmens, Miyeko Mana, Lorenzo de Lichtervelde, Joram D Mul, Diederik van de Peut, Maxime Devos, Nicolas Simonis, Muhammed A Yildirim, Murat Cokol, Huey-Ling Kao, Anne-Sophie de Smet, Haidong Wang, Anne-Lore Schlaitz, Tong Hao, Stuart Milstein, Changyu Fan, Mike Tipsword, Kevin Drew, Matilde Galli, Kahn Rhrissorrakrai, David Drechsel, Daphne Koller, Frederick P Roth, Lilia M Iakoucheva, A Keith Dunker, Richard Bonneau, Kristin C Gunsalus, David E Hill, Fabio Piano, Jan Tavernier, Sander van den Heuvel, Anthony A Hyman, Marc Vidal
Affiliations
- PMID: 18692475
- PMCID: PMC2596478
- DOI: 10.1016/j.cell.2008.07.009
A protein domain-based interactome network for C. elegans early embryogenesis
Mike Boxem et al. Cell. 2008.
Erratum in
- Cell. 2012 Dec 21;151(7):1633
Abstract
Many protein-protein interactions are mediated through independently folding modular domains. Proteome-wide efforts to model protein-protein interaction or "interactome" networks have largely ignored this modular organization of proteins. We developed an experimental strategy to efficiently identify interaction domains and generated a domain-based interactome network for proteins involved in C. elegans early-embryonic cell divisions. Minimal interacting regions were identified for over 200 proteins, providing important information on their domain organization. Furthermore, our approach increased the sensitivity of the two-hybrid system, resulting in a more complete interactome network. This interactome modeling strategy revealed insights into C. elegans centrosome function and is applicable to other biological processes in this and other organisms.
Figures
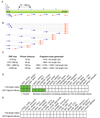
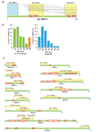
Figure 1. Strategy for generating the AD-Fragment library and effect on Y2H sensitivity and specificity
(A) Primer placement. Primers are designed to start within a 55 bp window surrounding the ideal start positions (lines above ORF). (B) Fragments generated by combining primers. (C) Distances in between primers and fragment sizes produced for ORFs of the indicated lengths. (D,E) Literature derived interactions and random protein pairs tested as full-length fusions (results from Venkatesan et al. personal communication) and using an AD-Fragment library. Green boxes indicate detection of an interaction. Protein names correspond to Entrez names.
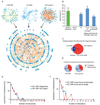
Figure 2. Properties of the Y2H protein-protein interaction network
(A) Network graph of the protein-protein interactions between early embryogenesis proteins, compiled from data in the most recent release of the worm interactome (CCSB-WI8), and from the AD-cDNA and AD-Fragment screens described here. (B) Retest rate of interactions in MAPPIT. Green bar: interactions derived from literature (results from Simonis et. al. personal communication). Random protein pairs did not interact. Blue bars: retest of 355 interactions described here, split into: (1) all 355 interactions, (2) those found as full-length fusions (124 interactions), and (3) those found as truncated fusions only (225 interactions). Error bars correspond to binomial standard error. (C) Overlap between AD-cDNA and AD-Fragment library derived interactions within the early embryogenesis protein space. (D) Fraction of interactions found as full-length fusions in AD-cDNA and AD-Fragment library screens. (E) Comparison of connectivity of bait and prey proteins. (F) Comparison of connectivity of prey proteins that were found as full-length at least once, with those that were never found as full-length.
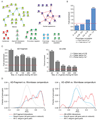
Figure 3. Enrichment in similar phenotypes, GO terms, and mRNA expression profiles for interacting protein pairs
(A) Examples of interactions between proteins assigned to the same functional class based on their RNAi phenotypes. Red lines: new Y2H interactions. Blue lines: known Y2H interactions re-identified. Blue dotted lines: known Y2H interactions not found. (B) Enrichment in phenotypic correlation for interacting protein pairs relative to average value of all possible protein pairs in the interaction network. (C) Enrichment in shared GO terms at different levels of specificity. (D) Pearson correlation coefficients (PCCs) for the mRNAs corresponding to each pair of proteins in the interaction data sets (red lines), the protein space searched (blue lines), and the entire worm genome (dotted grey lines). Early embryogenesis genes already have highly similar expression profiles compared to the entire worm genome, hence no further enrichment can be observed for interactions derived from the AD-Fragment library (left panel).
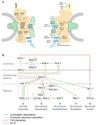
Figure 4. Y2H results of nuclear pore complex (NPC) and centrosome screens
(A) Schematic drawing of NPC. Shown are nuclear membrane (grey) with membrane rings (green), inner and outer scaffold rings (orange), FG nucleoporins (green), cytoplasmic tendrils (yellow), and nuclear basket (blue). Left: approximate localization of mammalian proteins within the NPC. C. elegans homologs of proteins in black were used as baits in our screens. Right: Interactions found between C. elegans NPC and import/export machinery proteins. (B) Diagram of centrosome assembly pathway. Green arrows represent localization dependencies, dotted blue lines previously described binary interactions, red lines Y2H interactions discovered here, and dotted boxes co-IP complexes.
Figure 5. Identification and validation of minimal regions required for interaction (MRIs)
(A) Example of identification of an MRI. The AD-Fragment library was screened with full length DB∷RAN- 1 and DB::IMB-4. Grey lines indicate protein fragments of NPP-9 that interacted with RAN-1 or IMB-4. (B) Sizes of MRIs identified in the AD-Fragment library screens expressed as percentage of corresponding full-length protein and absolute amino acids. (C) MRIs identified in proteins involved in centrosome assembly. Green bars represent full-length proteins. Yellow bars represent regions of the full-length protein required for interaction with the indicated binding partner (e.g. the N-terminal region of TPXL-1 is required for binding to AIR-1). Pfam-A domain signatures are drawn as red boxes. CC = coiled-coil prediction. The region of RSA-2 that mediates binding to SPD-5 was further refined manually (not shown).
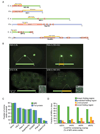
Figure 6. Comparison of MRIs with computational domain predictions
(A) Three cases where interacting regions differ between C. elegans and the orthologous proteins in human. (B) Localization of GFP fusions of full-length RSA-2 and SAS-5 and their MRIs required for binding to SPD-5 and SAS-6, respectively. (C) Fraction of amino acids of MRIs and the corresponding full proteins that are covered by computationally predicted domains of the indicated types. (D) Fraction of MRIs classified as ‘known folding region,’ ‘predicted folding region,’ ‘unstructured,’ or ‘novel folding region,’ based on overlap with computational predictions.
Similar articles
- Information flow analysis of interactome networks.
Missiuro PV, Liu K, Zou L, Ross BC, Zhao G, Liu JS, Ge H. Missiuro PV, et al. PLoS Comput Biol. 2009 Apr;5(4):e1000350. doi: 10.1371/journal.pcbi.1000350. Epub 2009 Apr 10. PLoS Comput Biol. 2009. PMID: 19503817 Free PMC article. - Physically asymmetric division of the C. elegans zygote ensures invariably successful embryogenesis.
Jankele R, Jelier R, Gönczy P. Jankele R, et al. Elife. 2021 Feb 23;10:e61714. doi: 10.7554/eLife.61714. Elife. 2021. PMID: 33620314 Free PMC article. - Identification of human protein interaction domains using an ORFeome-based yeast two-hybrid fragment library.
Waaijers S, Koorman T, Kerver J, Boxem M. Waaijers S, et al. J Proteome Res. 2013 Jul 5;12(7):3181-92. doi: 10.1021/pr400047p. Epub 2013 Jun 17. J Proteome Res. 2013. PMID: 23718855 - [Specification of cell destiny in early Caenorhabditis elegans embryo].
Schierenberg E. Schierenberg E. Naturwissenschaften. 1997 Feb;84(2):55-64. doi: 10.1007/s001140050349. Naturwissenschaften. 1997. PMID: 9121590 Review. German. - Cell cycle timing regulation during asynchronous divisions of the early C. elegans embryo.
Tavernier N, Labbé JC, Pintard L. Tavernier N, et al. Exp Cell Res. 2015 Oct 1;337(2):243-8. doi: 10.1016/j.yexcr.2015.07.022. Epub 2015 Jul 23. Exp Cell Res. 2015. PMID: 26213213 Review.
Cited by
- Cyclin CYB-3 controls both S-phase and mitosis and is asymmetrically distributed in the early C. elegans embryo.
Michael WM. Michael WM. Development. 2016 Sep 1;143(17):3119-27. doi: 10.1242/dev.141226. Development. 2016. PMID: 27578178 Free PMC article. - Bacterial protein networks: properties and functions.
Typas A, Sourjik V. Typas A, et al. Nat Rev Microbiol. 2015 Sep;13(9):559-72. doi: 10.1038/nrmicro3508. Epub 2015 Aug 10. Nat Rev Microbiol. 2015. PMID: 26256789 Review. - SYNZIP protein interaction toolbox: in vitro and in vivo specifications of heterospecific coiled-coil interaction domains.
Thompson KE, Bashor CJ, Lim WA, Keating AE. Thompson KE, et al. ACS Synth Biol. 2012 Apr 20;1(4):118-29. doi: 10.1021/sb200015u. ACS Synth Biol. 2012. PMID: 22558529 Free PMC article. - Multisite Phosphorylation of NuMA-Related LIN-5 Controls Mitotic Spindle Positioning in C. elegans.
Portegijs V, Fielmich LE, Galli M, Schmidt R, Muñoz J, van Mourik T, Akhmanova A, Heck AJ, Boxem M, van den Heuvel S. Portegijs V, et al. PLoS Genet. 2016 Oct 6;12(10):e1006291. doi: 10.1371/journal.pgen.1006291. eCollection 2016 Oct. PLoS Genet. 2016. PMID: 27711157 Free PMC article. - aPKC phosphorylates NuMA-related LIN-5 to position the mitotic spindle during asymmetric division.
Galli M, Muñoz J, Portegijs V, Boxem M, Grill SW, Heck AJ, van den Heuvel S. Galli M, et al. Nat Cell Biol. 2011 Aug 21;13(9):1132-8. doi: 10.1038/ncb2315. Nat Cell Biol. 2011. PMID: 21857670
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. - PubMed
- Bornberg-Bauer E, Beaussart F, Kummerfeld SK, Teichmann SA, Weiner J., 3rd The evolution of domain arrangements in proteins and interaction networks. Cell Mol Life Sci. 2005;62:435–445. - PubMed
- Chivian D, Kim DE, Malmstrom L, Bradley P, Robertson T, Murphy P, Strauss CE, Bonneau R, Rohl CA, Baker D. Automated prediction of CASP-5 structures using the Robetta server. Proteins. 2003;53 Suppl 6:524–533. - PubMed
- Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7:815–829. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21CA113711/CA/NCI NIH HHS/United States
- R01 HG001715-06/HG/NHGRI NIH HHS/United States
- R21 CA113711-01A2/CA/NCI NIH HHS/United States
- R33CA81658/CA/NCI NIH HHS/United States
- R33 CA105405-02/CA/NCI NIH HHS/United States
- R01HG001715/HG/NHGRI NIH HHS/United States
- R33 CA081658-04/CA/NCI NIH HHS/United States
- R21RR023114/RR/NCRR NIH HHS/United States
- P01 CA095281-01A10004/CA/NCI NIH HHS/United States
- U54 CA011295/CA/NCI NIH HHS/United States
- P01 CA095281/CA/NCI NIH HHS/United States
- U54 CA112952-01/CA/NCI NIH HHS/United States
- U54 CA112952-02/CA/NCI NIH HHS/United States
- R21 RR023114-02/RR/NCRR NIH HHS/United States
- R21 RR023114/RR/NCRR NIH HHS/United States
- R21 RR023114-01/RR/NCRR NIH HHS/United States
- R01 HG001715-07/HG/NHGRI NIH HHS/United States
- CA95281/CA/NCI NIH HHS/United States
- U54 CA112952-03/CA/NCI NIH HHS/United States
- R33 CA105405-03/CA/NCI NIH HHS/United States
- R01 HG001715/HG/NHGRI NIH HHS/United States
- R33CA105405/CA/NCI NIH HHS/United States
- R01 HG001715-09/HG/NHGRI NIH HHS/United States
- R33 CA105405/CA/NCI NIH HHS/United States
- R01 HG001715-10/HG/NHGRI NIH HHS/United States
- U54 CA112952/CA/NCI NIH HHS/United States
- R21 CA113711/CA/NCI NIH HHS/United States
- R33 CA105405-01/CA/NCI NIH HHS/United States
- R01 HG001715-07S1/HG/NHGRI NIH HHS/United States
- R01 HG001715-08/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases