Transcription-independent functions of MYC: regulation of translation and DNA replication - PubMed (original) (raw)
Review
Transcription-independent functions of MYC: regulation of translation and DNA replication
Michael D Cole et al. Nat Rev Mol Cell Biol. 2008 Oct.
Abstract
MYC is a potent oncogene that drives unrestrained cell growth and proliferation. Shortly after its discovery as an oncogene, the MYC protein was recognized as a sequence-specific transcription factor. Since that time, MYC oncogene research has focused on the mechanism of MYC-induced transcription and on the identification of MYC transcriptional target genes. Recently, MYC was shown to control protein expression through mRNA translation and to directly regulate DNA replication, thus initiating exciting new areas of oncogene research.
Figures
Figure 1. The conserved regions of MYC
The three MYC proteins (MYC, MYCN and MYCL) are encoded by separate genes with distinct developmental regulation, but all three have been directly implicated in cancer. The N terminus of MYC contains the transactivation domain and the C terminus contains the DNA-binding domain. The MYC boxes I, II, III and IV are indicated in red. The basic helix-loop- helix/Leu zipper (bHLH/LZ) domain is indicated in green. MYC box II (MBII) has been shown to have a crucial role in most of the biological activities of MYC. Note that MBIV is not a component of the minimal DNA-binding domain but does influence DNA binding in vivo.
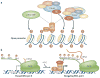
Figure 2. Mechanisms of MYC-induced transcription
a | MYC recruits histone acetyltransferases, which promote localized modification of chromatin through acetylation of nucleosomes. MYC-associated factor-X (MAX) dimerizes with MYC to create a functional DNA-binding complex that preferentially recognizes the consensus sequence CACGTG. The MYC transactivation domain recruits acetyltransferase complexes that contain transformation/transcription-domain-associated protein (TRRAP) and either general control of amino-acid-synthesis protein-5 (GCN5) or TIP60, which preferentially acetylate histones H3 or H4, respectively. MYC can also recruit p300/CReB-binding protein (CBP). These complexes alter the acetylation of nucleosomes around MYC target genes, and these acetylation marks might also recruit other nuclear factors that influence transcription, such as the SWI/SNF chromatin-remodelling complex (not shown). b | MYC recruits basal transcription factors and promotes the clearance of promoters through RNA polymerase (pol) II. RNA pol II is frequently paused on promoters after phosphorylation of Ser5 on the RNA pol II C-terminal domain (CTD) and synthesis of a short (20–40 base) segment of mRNA. The MYC protein can promote a paused RNA pol to continue transcription of the mRNA by recruiting the P-TeFb (positive transcription-elongation factor-b) complex, which phosphorylates the CTD on Ser2 and promotes transcriptional elongation.
Figure 3. Mechanism of MYC-induced mrNa cap methylation
MYC promotes recruitment of the transcription factor TFIIH kinase to promoters and RNA polymerase (pol) II phosphorylation. Increased RNA pol II phosphorylation increases cap RNA methyltransferase (RNMT) recruitment and/or activity, which correlates with MYC-dependent mRNA cap methylation. At direct MYC target genes (left), TFIIH enhances the recruitment or activity of the cap RNMT to increase the fraction of cap methylated mRNA. Direct binding of MYC also stimulates a modest increase in transcription through TFIIH or acetyltransferases (see FIG. 2). At other promoters (right), MYC stimulates mRNA cap methylation through TFIIH stimulation by the MYC transactivation domain and through the subsequent recruitment or activation of RNMT by C-terminal domain phosphorylation. In doing so, MYC functions as a transcription-independent factor. Activation of direct targets is MYC-associated factor-X (MAX)-dependent (left), whereas activation of transcription-independent targets is MAX-independent (right).
Similar articles
- Myc up-regulates formation of the mRNA methyl cap.
Cowling VH. Cowling VH. Biochem Soc Trans. 2010 Dec;38(6):1598-601. doi: 10.1042/BST0381598. Biochem Soc Trans. 2010. PMID: 21118133 - MYC as a regulator of ribosome biogenesis and protein synthesis.
van Riggelen J, Yetil A, Felsher DW. van Riggelen J, et al. Nat Rev Cancer. 2010 Apr;10(4):301-9. doi: 10.1038/nrc2819. Nat Rev Cancer. 2010. PMID: 20332779 Review. - c-Myc deregulation induces mRNA capping enzyme dependency.
Lombardi O, Varshney D, Phillips NM, Cowling VH. Lombardi O, et al. Oncotarget. 2016 Dec 13;7(50):82273-82288. doi: 10.18632/oncotarget.12701. Oncotarget. 2016. PMID: 27756891 Free PMC article. - MYC Mediates mRNA Cap Methylation of Canonical Wnt/β-Catenin Signaling Transcripts By Recruiting CDK7 and RNA Methyltransferase.
Posternak V, Ung MH, Cheng C, Cole MD. Posternak V, et al. Mol Cancer Res. 2017 Feb;15(2):213-224. doi: 10.1158/1541-7786.MCR-16-0247. Epub 2016 Nov 29. Mol Cancer Res. 2017. PMID: 27899423 Free PMC article. - Target gene-independent functions of MYC oncoproteins.
Baluapuri A, Wolf E, Eilers M. Baluapuri A, et al. Nat Rev Mol Cell Biol. 2020 May;21(5):255-267. doi: 10.1038/s41580-020-0215-2. Epub 2020 Feb 18. Nat Rev Mol Cell Biol. 2020. PMID: 32071436 Free PMC article. Review.
Cited by
- S146L in MYC is a context-dependent activating substitution in cancer development.
Hinds JW, Feris EJ, Wilkins OM, Deary LT, Wang X, Cole MD. Hinds JW, et al. PLoS One. 2022 Aug 26;17(8):e0272771. doi: 10.1371/journal.pone.0272771. eCollection 2022. PLoS One. 2022. PMID: 36018850 Free PMC article. - Extending the functions of the homeotic transcription factor Cdx2 in the digestive system through nontranscriptional activities.
Freund JN, Duluc I, Reimund JM, Gross I, Domon-Dell C. Freund JN, et al. World J Gastroenterol. 2015 Feb 7;21(5):1436-43. doi: 10.3748/wjg.v21.i5.1436. World J Gastroenterol. 2015. PMID: 25663763 Free PMC article. Review. - MTBP and MYC: A Dynamic Duo in Proliferation, Cancer, and Aging.
Grieb BC, Eischen CM. Grieb BC, et al. Biology (Basel). 2022 Jun 8;11(6):881. doi: 10.3390/biology11060881. Biology (Basel). 2022. PMID: 35741402 Free PMC article. Review. - Deubiquitinase USP13 maintains glioblastoma stem cells by antagonizing FBXL14-mediated Myc ubiquitination.
Fang X, Zhou W, Wu Q, Huang Z, Shi Y, Yang K, Chen C, Xie Q, Mack SC, Wang X, Carcaboso AM, Sloan AE, Ouyang G, McLendon RE, Bian XW, Rich JN, Bao S. Fang X, et al. J Exp Med. 2017 Jan;214(1):245-267. doi: 10.1084/jem.20151673. Epub 2016 Dec 6. J Exp Med. 2017. PMID: 27923907 Free PMC article. - The opposing transcriptional functions of Sin3a and c-Myc are required to maintain tissue homeostasis.
Nascimento EM, Cox CL, MacArthur S, Hussain S, Trotter M, Blanco S, Suraj M, Nichols J, Kübler B, Benitah SA, Hendrich B, Odom DT, Frye M. Nascimento EM, et al. Nat Cell Biol. 2011 Nov 20;13(12):1395-405. doi: 10.1038/ncb2385. Nat Cell Biol. 2011. PMID: 22101514 Free PMC article.
References
- Oster SK, Ho CS, Soucie EL, Penn LZ. The myc oncogene: MarvelouslY Complex. Adv Cancer Res. 2002;84:81–154. - PubMed
- Pirity M, Blanck JK, Schreiber-Agus N. Lessons learned from Myc/Max/Mad knockout mice. Curr Top Microbiol Immunol. 2006;302:205–234. - PubMed
- Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. - PubMed
- Dang CV, et al. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. - PubMed
- Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources