Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis - PubMed (original) (raw)
Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis
Pauline E Jullien et al. PLoS Biol. 2008.
Abstract
Parental genomic imprinting causes preferential expression of one of the two parental alleles. In mammals, differential sex-dependent deposition of silencing DNA methylation marks during gametogenesis initiates a new cycle of imprinting. Parental genomic imprinting has been detected in plants and relies on DNA methylation by the methyltransferase MET1. However, in contrast to mammals, plant imprints are created by differential removal of silencing marks during gametogenesis. In Arabidopsis, DNA demethylation is mediated by the DNA glycosylase DEMETER (DME) causing activation of imprinted genes at the end of female gametogenesis. On the basis of genetic interactions, we show that in addition to DME, the plant homologs of the human Retinoblastoma (Rb) and its binding partner RbAp48 are required for the activation of the imprinted genes FIS2 and FWA. This Rb-dependent activation is mediated by direct transcriptional repression of MET1 during female gametogenesis. We have thus identified a new mechanism required for imprinting establishment, outlining a new role for the Retinoblastoma pathway, which may be conserved in mammals.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
Figure 1. RBR1 Interacts with MSI1
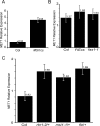
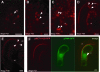
(A) Pull-down assay testing for interaction between Arabidopsis MSI1 and RBR1. The full-length RBR1 protein, labeled with [35S] methionine, was incubated with GST-MSI1 protein bound to agarose beads (GST MSI). As a negative control, the labeled protein was incubated with GST only, bound to agarose beads (GST). Input indicates the labeled protein used in the binding assays. Immunodetection of GST and GST-MSI1 were performed using anti-GST antibody. (B–H) BiFC analyses showing in vivo interaction between RBR1 and MSI1 proteins. Fluorescence is observed in nuclei following YFP reconstitution between YN-MSI1 and YC-RBR1 (B) The inset represents the detail of an individual nucleus. In comparison, either YN with YC-RBR1 (F) or YN-MSI1 with YC (G) serving as negative controls display no fluorescence. (C–E) Truncation of the RBR1 protein showing that only the RB-A domain is required for the interaction with MSI1. Localization was determined in leaf epidermis of N. benthamiana. YFP fluorescence from single confocal sections showing a fraction of the nuclei from all cells in the field was overlaid with Nomarsky differential interference contrast (DIC) images. Arrows point to nuclei expressing YFP fluorescence. Scale bars represent 20 μm. (H) Representation of the different RBR1 truncations tested for interaction with MSI1 in the assays shown in (A) to (G).
Figure 2. MSI1 Represses MET1 Expression
(A) Q-PCR analyses on RNAs from mature leaves show an increase of MET1 expression in MSI1cs in comparison to wild-type Columbia. The RQ value corresponds to the average of five independent biological replicates. (B) Q-PCR analyses on RNAs from FIEcs and fas1 mature leaves. (C) Q-PCR analyses showing an increase of MET1 expression in rbr1–1/+, msi1–1/+, and fie/+ ovules at 1.5 d after emasculation (DAE) in comparison to wild-type Col (B and C). The RQ value corresponds to the average of three independent biological replicates. (A–C) Error bars represent the standard error between the biological replicates. The RQ value is represented on the top of each bar. ACT11 was used as endogenous control for (A) and (B), GAPC for (C).
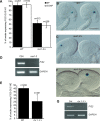
Figure 3. MSI1 and RBR1 Bind to the MET1 Promoter
(A) Schematic diagram of the MET1 locus representing the fragments (black rectangles) analyzed by PCR after ChIP. White boxes represent the 5′ UTR, and the gray arrow corresponds to the first amino acid of exon 1 of MET1. A putative E2F binding site is represented by a black dot. (B) ChIP analysis using antibodies specific for MSI1 (MSI1 Ab) and RBR1 (RBR1 Ab) proteins. Nuclear extracts were prepared from wild-type Columbia buds after cross-linking. The first lane represents the input DNA. Control IgG is used as a negative control, while an antibody against histone 3 (H3) is used as a positive control. (C) Absolute quantification of the ChIP using Q-PCR for the fragment 2. Error bars represent the standard deviation of two independent PCR reactions.
Figure 4. Expression the p_MSI1_-MSI1-mRFP1 Fusion Protein during Female Gametophyte Development
Confocal images from transgenic plants expressing the MSI1-mRFP fusion protein under the control of its native promoter. MSI1-mRFP is expressed in nuclei of all the cells of the ovule integuments (oi). (A) Ovule with the functional megaspore (fm), where MSI1-mRFP accumulates in the nucleus. (B) Four-nucleate–stage (FG4) ovule. Only two of the four nuclei are visible. (C) Eight-nucleate–stage (FG8) ovule. (D) Mature female gametophyte. cc, central cell; ec, egg cell; nu, nucellus; pn, polar nuclei; sy, synergids. Scale bars represent 10 μm for (A) and 20 μm for (B–D).
Figure 5. Expression the p_MET1_-H2B-RFP Reporter during Female Gametogenesis
Confocal images from transgenic plants expressing the p_MET1_-H2B-RFP construct. (A) p_MET1_-H2B-RFP is expressed in the functional megaspore (fm) as well as in the ovule integument (oi). (B) Two-nucleate–stage (FG2) ovule. (C) Four-nucleate–stage (FG4) ovule. (D) Eight-nucleate–stage (FG8) ovule; p_MET1_-H2B-RFP expression is restricted to the antipodal nuclei (an). (E) Mature female gametophyte. (F) Colocalization of p_MET1_-H2B-RFP with _FWA_-GFP showing an absence of RFP in the mature central cell (cc), 1DAE. ec, egg cell; sy, synergids. Stages of female gametophyte development are indicated according to Christensen et al. [68]. Arrowheads point to nuclei. Scale bars represent 10 μm.
Figure 6. Control of MET1 Expression by MSI1 and RBR1
(A) p_MET1_-H2B-RFP in wild-type mature ovules, the expression is restricted to the antipodal cells (an) and ovule integuments (oi). (B) p_MET1_-H2B-RFP in rbr1–1 ovules, ectopic RFP expression is observed in the central cell (cc) and egg cell (ec). (C) p_MET1_-H2B-RFP in msi1–1 ovule, ectopic RFP expression is observed in the central cell (cc) and egg cell (ec). Scale bar represent 10 μm. (D) Percentage of ovules expressing p_MET1_:H2B-RFP in the central cell in wild type (WT), rbr1–1/+, msi1–1/+, and fie/+ plants. Arrowheads point to nuclei. Error bars represent the standard deviation. The n number is represented on the top of each bar.
Figure 7. Control of FIS2 Expression by MSI1 and RBR1
(A) Percentage of ovules/seeds expressing GUS in plants homozygous (HO) for FIS2-GUS in wild-type (WT) and msi1–2/+ backgrounds. The percentage of ovules/seeds is represented before pollination (BP) and 3 d after pollination (3DAP). Error bars represent the standard deviation. The n number is represented on the top of each bar. (B and C) Photography illustrating FIS2-GUS expression in the central cell of wild-type (B) and msi1–2/+ (C) ovules. Before pollination, it is not possible to distinguish on morphological bases the gametes carrying the wild-type or the msi1–2 allele. Scale bars represent 50 μm. (D) RT-PCR on RNAs from seeds that inherited msi1–2 maternally selected on the basis of the overexpression of the fluorescent marker KS117 [28] (5 DAP). GAPDH is used as a control. (E) Percentage of ovules expressing GUS in plants hemizygous (HE) for FIS2-GUS in wild-type and rbr1–1/+ backgrounds. Error bars represent the standard deviation. The n number is represented on the top of each bar. (F) FIS2-GUS expression in the central cell of rbr1–1/+ ovules before pollination. Before pollination, it is not possible to distinguish on morphological bases the gametes carrying the wild-type or the rbr1–1 allele. Scale bars represent 50 μm. (G) RT-PCR on RNAs from _rbr1–1_–selected ovules showing no FIS2 expression in comparison to wild-type ovules. Selection of rbr1–1 ovules was based on the lack of fertilization and seed development at 3 DAP in contrast to the wild-type ovules that were fertilized. GAPDH is used as a control.
Figure 8. RBR1 and MSI1 Control Maternal FWA Expression but Not Maternal MEA Expression
(A) Percentage of ovules expressing GFP or GUS activity from plants homozygous for FWA-GFP and MEA-GUS in wild-type (black bars), msi1–2/+ (grey bars), and rbr1–1/+ (white bars) backgrounds. Error bars represent the standard deviation. The n number is represented on the top of each bar. (B–D) FWA-GFP fluorescence in the central cell of wild-type (B), msi1–2/+ (C), and rbr1–1/+ (D) ovules. (E–G) MEA-GUS staining in the central cell of WT (E), msi1–2/+ (F), and rbr1–1/+ (G) ovules. Scale bars represent 50 μm.
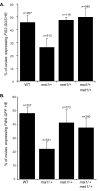
Figure 9. MSI1 Activation of FIS2 and FWA Is Mediated through MET1
Percentage of ovules expressing GFP or GUS activity from plants hemizygous (HE) for FIS2-GUS (A) and FWA-GFP (B) constructs in wild-type, msi1–2/+, met1–3/+, and msi1–3/+ met1–3/+ backgrounds. Error bars represent the standard deviation. The n number is represented on the top of each bar.
Figure 10. Model for MSI1/RBR1 Regulation of FIS2 and FWA Maternal Expression
The MSI1/RBR1 complex represses the expression of MET1 during female gametogenesis. As a result, the silencing DNA methylation marks (pink lollipops) are gradually lost during the female gamete nuclei divisions. In the central cell, DME removes the residual marks on imprinted genes such as FIS2 and FWA, resulting in their transcriptional activation. The active status is conserved on the maternal allele during endosperm development. During male gametogenesis, MET1 is expressed (Figure S7) and maintains the repression on the FIS2 and FWA paternal allele. The paternal copy remains silent during endosperm development.
Comment in
- Retinoblastoma makes its mark on imprinting in plants.
Costa LM, Gutierrez-Marcos JF. Costa LM, et al. PLoS Biol. 2008 Aug 26;6(8):e212. doi: 10.1371/journal.pbio.0060212. PLoS Biol. 2008. PMID: 18752352 Free PMC article.
Similar articles
- One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation.
Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T. Kinoshita T, et al. Science. 2004 Jan 23;303(5657):521-3. doi: 10.1126/science.1089835. Epub 2003 Nov 20. Science. 2004. PMID: 14631047 - Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting.
Jullien PE, Kinoshita T, Ohad N, Berger F. Jullien PE, et al. Plant Cell. 2006 Jun;18(6):1360-72. doi: 10.1105/tpc.106.041178. Epub 2006 Apr 28. Plant Cell. 2006. PMID: 16648367 Free PMC article. - HMG domain containing SSRP1 is required for DNA demethylation and genomic imprinting in Arabidopsis.
Ikeda Y, Kinoshita Y, Susaki D, Ikeda Y, Iwano M, Takayama S, Higashiyama T, Kakutani T, Kinoshita T. Ikeda Y, et al. Dev Cell. 2011 Sep 13;21(3):589-96. doi: 10.1016/j.devcel.2011.08.013. Dev Cell. 2011. PMID: 21920319 - Gamete-specific epigenetic mechanisms shape genomic imprinting.
Jullien PE, Berger F. Jullien PE, et al. Curr Opin Plant Biol. 2009 Oct;12(5):637-42. doi: 10.1016/j.pbi.2009.07.004. Epub 2009 Aug 24. Curr Opin Plant Biol. 2009. PMID: 19709923 Review. - Imprinting in plants and its underlying mechanisms.
Zhang H, Chaudhury A, Wu X. Zhang H, et al. J Genet Genomics. 2013 May 20;40(5):239-47. doi: 10.1016/j.jgg.2013.04.003. Epub 2013 Apr 20. J Genet Genomics. 2013. PMID: 23706299 Review.
Cited by
- RNA-directed DNA methylation regulates parental genomic imprinting at several loci in Arabidopsis.
Vu TM, Nakamura M, Calarco JP, Susaki D, Lim PQ, Kinoshita T, Higashiyama T, Martienssen RA, Berger F. Vu TM, et al. Development. 2013 Jul;140(14):2953-60. doi: 10.1242/dev.092981. Epub 2013 Jun 12. Development. 2013. PMID: 23760956 Free PMC article. - Genome-wide Identification, Expression, and Functional Analysis of MdMSI Genes in Apples (Malus domestica Borkh.).
Wang D, Wang X, Zhang C, Yang K, Wang X, Cui J, Liu D, You C. Wang D, et al. Front Genet. 2022 Mar 3;13:846321. doi: 10.3389/fgene.2022.846321. eCollection 2022. Front Genet. 2022. PMID: 35309144 Free PMC article. - Looking at plant cell cycle from the chromatin window.
Desvoyes B, Fernández-Marcos M, Sequeira-Mendes J, Otero S, Vergara Z, Gutierrez C. Desvoyes B, et al. Front Plant Sci. 2014 Jul 25;5:369. doi: 10.3389/fpls.2014.00369. eCollection 2014. Front Plant Sci. 2014. PMID: 25120553 Free PMC article. Review. - Loss of the DNA methyltransferase MET1 Induces H3K9 hypermethylation at PcG target genes and redistribution of H3K27 trimethylation to transposons in Arabidopsis thaliana.
Deleris A, Stroud H, Bernatavichute Y, Johnson E, Klein G, Schubert D, Jacobsen SE. Deleris A, et al. PLoS Genet. 2012;8(11):e1003062. doi: 10.1371/journal.pgen.1003062. Epub 2012 Nov 29. PLoS Genet. 2012. PMID: 23209430 Free PMC article. - DNA METHYLTRANSFERASE 1 is involved in (m)CG and (m)CCG DNA methylation and is essential for sporophyte development in Physcomitrella patens.
Yaari R, Noy-Malka C, Wiedemann G, Auerbach Gershovitz N, Reski R, Katz A, Ohad N. Yaari R, et al. Plant Mol Biol. 2015 Jul;88(4-5):387-400. doi: 10.1007/s11103-015-0328-8. Epub 2015 May 6. Plant Mol Biol. 2015. PMID: 25944663
References
- Feil R, Berger F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 2007;23:192–199. - PubMed
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. - PubMed
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, et al. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. - PubMed
- Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. - PubMed
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet 14 Spec No. 2005;1:R47–58. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases