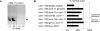OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development - PubMed (original) (raw)
OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development
Hidetoshi Komatsu et al. PLoS Biol. 2008.
Abstract
Notch signaling is critical for cell fate decisions during development. Caenorhabditis elegans and vertebrate Notch ligands are more diverse than classical Drosophila Notch ligands, suggesting possible functional complexities. Here, we describe a developmental role in Notch signaling for OSM-11, which has been previously implicated in defecation and osmotic resistance in C. elegans. We find that complete loss of OSM-11 causes defects in vulval precursor cell (VPC) fate specification during vulval development consistent with decreased Notch signaling. OSM-11 is a secreted, diffusible protein that, like previously described C. elegans Delta, Serrate, and LAG-2 (DSL) ligands, can interact with the lineage defective-12 (LIN-12) Notch receptor extracellular domain. Additionally, OSM-11 and similar C. elegans proteins share a common motif with Notch ligands from other species in a sequence defined here as the Delta and OSM-11 (DOS) motif. osm-11 loss-of-function defects in vulval development are exacerbated by loss of other DOS-motif genes or by loss of the Notch ligand DSL-1, suggesting that DOS-motif and DSL proteins act together to activate Notch signaling in vivo. The mammalian DOS-motif protein Deltalike1 (DLK1) can substitute for OSM-11 in C. elegans development, suggesting that DOS-motif function is conserved across species. We hypothesize that C. elegans OSM-11 and homologous proteins act as coactivators for Notch receptors, allowing precise regulation of Notch receptor signaling in developmental programs in both vertebrates and invertebrates.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
Figure 1. OSM-11 Is Required for Normal Development
(A) Thirty-one percent of osm-11(lf) adult animals had overtly normal vulva and did not retain eggs resembling control animals. (B) Fifty-seven percent of osm-11(lf) animals inappropriately retained eggs and/or had a single misshapen or protruding vulva (15% and 42%, respectively). (C) Twelve percent of osm-11(lf) animals had an extra protrusion near the normally positioned vulva. (D) osm-11(lf) animals had defects in head morphology at low frequency (arrowhead). (E) Two thirds of osm-11(lf) animals had a ventral protrusion behind the anus (arrowhead). n > 100 animals were scored.
Figure 2. osm-11 Encodes a Protein with a Conserved Motif Found in Notch Ligands
(A) Top: OSM-11 genomic structure. The signal peptide is shaded black, and putative O-linked glycosylation sites are indicated by vertical lines. The DOS motif is shaded blue; it overlaps the previously defined osmotic stress resistant (OSR) motif [52]. osm-11(rt142) removes all coding sequence after the signal peptide; osm-11(rt68) converts W177 to a premature stop codon. Bottom: the DOS motif-containing sequences from C. elegans OSM-11, OSM-7, DOS-1, DOS-2, and DOS-3 are aligned above the DOS motif consensus and the cEGF-1 consensus [53]. DOS-motif regions from mouse proteins and known Drosophila Notch ligands are aligned under the cEGF-1 consensus. DOS-motif amino acids are shaded blue and previously described EGF repeats are boxed. Asterisks (*) indicate cysteines in the conserved EGF-motif that are not found in the C. elegans DOS proteins. The DOS motif consensus is: C-X(3)-C-X(3,8)-C-X(2,5)-C-[KVER]-C-X(10,12)-C-X(1,3)-P-X(6,9)-C-X(1,4)-W-X(1,4)-C. In the DOS motif consensus, b represents K, V, E, or R, and the dash (-) indicates possible positions for proline in the DOS motif. In the cEGF-1 consensus, s represents a small amino acid [53]. (B) The position of the DOS motif in known or predicted C. elegans, Drosophila, and vertebrate Notch ligands. The DOS motif overlaps with the first two EGF repeats of canonical Notch ligands and may define a unique subset of EGF repeats. The noncanonical Notch ligands DNER [40], F3/contactin [95], and MAGP [–40] do not contain a DOS motif (unpublished data). (C) Similarity between DOS motifs, the first and second EGF repeats, and the third and fourth EGF repeats of Notch ligands. As noted by Lissemore and Starmer [27], the first and second EGF repeats differ from the third and fourth EGF repeats. DOS-3 was not included in this alignment. Green indicates the DOS motif of proteins that lack canonical EGF repeats; blue indicates the first and second EGF repeats of Notch ligands; red indicates the third and fourth EGF repeats of Notch ligands; and magenta represents the C. elegans Notch ligands that lack DSL domains. See Materials and Methods for accession numbers and other details.
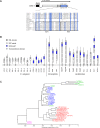
Figure 3. OSM-11 Loss Results in Cell Fate Specification Defects
(A) A simplified diagram of cell fate GFP marker expression in P5.p, P6.p, and P7.p. GFP expression is schematically shown in green. Note that equivalence group members P3.p, P4.p, and P8.p are not shown. In wild-type animals, primary (1°) cell fate markers are expressed in P6.p (top left), whereas secondary (2°) cell fate markers are normally expressed in P5.p and P7.p (top right). The first Pn.p division (by P5.p, P6.p, and P7.p) in mid-L3 larvae gives rise to Pn.px cells; the next divisions give rise to Pn.pxx cells in late-L3 larvae. Loss of Notch signaling does not stop 1° cell fate assumption by P6.p, but results in inappropriate adoption of 1° cell fates by P5.p, P7.p, and their descendents. Loss of EGF/Ras signaling results in adoption of the tertiary fate by P5.p and P7.p, and in some cases, P6.p, depending on the severity of the defect [96]. (B) Quantification of data from (C–E). p < 0.05 based on χ2 for each marker. (C) Ten percent of L3 osm-11(lf) animals (right) ectopically express the 1° cell fate marker egl-17p::gfp in P5.p or P7.p, which normally adapt the 2° fate (left). (D) Sixty-seven percent of L3 osm-11(lf) animals lack expression of the 2° cell fate marker lin-11p::gfp in descendants of P5.p and/or P7.p. (E) Thirty-five percent of L3 osm-11(lf) animals do not up-regulate expression of the 2° fate marker lip-1p::gfp in P5.p and/or P7.p. p < 0.05 based on χ2 for each marker. These data suggest osm-11(lf) animals have a loss of 2° cell fate specification consistent with loss of LIN-12 Notch signaling. In (C–E), arrowheads indicate the positions of P5.p, P6.p, and P7.p.
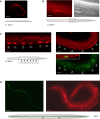
Figure 4. osm-11 Is Expressed in VPCs and Other Tissues
(A) OSM-11 expression in seam cells of L1 larvae detected using α-OSM-11 antisera. The seam cells on the right side of an L1 animal are in focus; the seam cells on the left side are visible and slightly out of focus. OSM-11 was not expressed in seam cells or hypoderm at other larval stages. (B) OSM-11 expression in the developing uterus of L4 larvae. Left, α-OSM-11 antisera staining; right, visible light image. (C) OSM-11 expression in vulval precursor cells (VPCs; arrowheads) in L3 larvae. The top panels show α-OSM-11 antisera staining of VPCs prior (top left) and immediately after (top right) cell fate specification as assessed by lip-1p::gfp expression. An overlay of α-OSM-11 staining and lip-1p::gfp expression shows that OSM-11 is concentrated on the apical surface of the VPCs (bottom right); this was confirmed using an ajm-1::gfp fusion (unpublished data). (D) OSM-11 expression in seam cells and spermatheca in adult animals. An osm-11p::gfp reporter gene containing unc-54 3′ UTR sequences is expressed in adult seam cells (left); α-OSM-11 antisera was used to confirm seam cell and spermatheca expression (right). No OSM-11 was detected in neurons of larvae or adult animals (unpublished data); embryonic expression was not characterized. In (A–D), the scale bar represents 10 μm.
Figure 5. osm-11 Encodes a Secreted Protein Required for Vulval Development
(A) Western blot of conditioned media from Drosophila S2 cells containing an OSM-11 cDNA expression construct or empty vector. OSM-11 was not detected in cell lysates (unpublished data). The molecular weight of mature OSM-11 was predicted at 18.9 kDa; the detected protein migrated at 20.7 kDa (arrowhead). OSM-11 may be O-linked glycosylated (see Figure 2). (B) Transgenic rescue of osm-11(lf) vulval defects. osm-11(lf) animals harboring transgenes with empty expression vectors were indistinguishable from nontransgenic osm-11(lf) animals (n = 129 animals, 5 transgenic lines) and were used as controls. Multiple transgenic lines were scored for all rescue experiments; data are reported as mean ± standard error of the mean (S.E.M.) In addition to a genomic osm-11 construct, expression of the osm-11 cDNA using the following promoters also significantly rescued osm-11(lf) vulval defects: osm-11p, hsp-16p (ubiquitous expression; 79% normal vulval; unpublished data), wrt-6p (hypodermal), osm-10p (sensory neurons), and glr-1p (nonoverlapping set of neurons vs. osm-10p). In addition, heterologous expression of mammalian DLK1 driven by the hsp-16 promoter also significantly rescued osm-11(lf) vulval phenotypes. n > 52 animals for each transgene, p < 0.05 by χ2.
Figure 6. osm-11 Normally Increases Notch Signaling during Vulval Development
(A and B) lin-12(lf) is epistatic to osm-11(lf). lin-12(lf) is the null allele n941; animals carrying this allele have a protruding vulva (pVul; [A]) that is distinct from the defective vulva seen in osm-11(lf) animals (see Figure 1). lin-12(lf);osm-11(lf) animals were indistinguishable from lin-12(lf) animals (B). (C and D) osm-11(lf) suppresses lin-12(csgf) at 15 °C. lin-12(csgf) is n137n460, a recessive cold-sensitive gain-of-function allele; animals carrying this mutation have multiple pseudovulvae (Muv; [C]). lin-12(csgf);osm-11(lf) animals were significantly less Muv (nonMuv) than lin-12(csgf) animals ([D]; p < 0.05). (E and F) _osm-11(lf)_ does not suppress _lin-12(gf)_. _lin-12(gf)_ is _n137_, a dominant gain-of-function allele that is ligand independent; animals carrying this mutation are Muv (E). _lin-12(gf);osm-11(lf)_ animals were indistinguishable from _lin-12(gf)_ animals (F). _n_ > 50 animals were scored for each genotype.
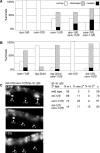
Figure 7. OSM-11 Acts Synergistically with DSL Ligands and Other DOS Proteins
In (A and B), phenotypes were scored as in Figure 1. (A) Genetic interactions between osm-11 and DOS-motif genes osm-7 and dos-1 (ZK507.4). dos-1(lf) and osm-7(lf) are both presumptive null alleles, and animals harboring these alleles had normal vulvas. dos-1(lf);osm-11(lf) and osm-7(lf);osm-11(lf) animals had significantly more severe defects than osm-11(lf) animals (p < 0.005, χ2 test). Mutant alleles of dos-2 (K10G6.2) and dos-3 (K02F3.7) are not currently available. (B) Genetic interactions between osm-11 and DSL-domain genes lag-2 and dsl-1. lag-2(dn) is the dominant negative allele sa37; dsl-1(lf) is ok810 and is a presumptive null allele. lag-2(dn) and dsl-1(lf) animals had few or no vulval defects. lag-2(dn);osm-11(lf) and dsl-1(lf);osm-11(lf) animals had significantly more-severe defects that osm-11(lf) animals (p < 0.005, χ2 test). (C) Vulval precursor cell (VPC) fate analysis for osm-11 and dsl-1. Arrowheads indicate the positions of P5.p, P6.p, and P7.p. Secondary (2°) cell fates were scored as in Figure 3 using lip-1p::GFP as illustrated (right). dsl-1;osm-11 double-mutant animals had significantly more severe 2° fate specification defects compared to either single mutant alone (p < 0.005 by χ2). n ≥ 48 for each genotype in all panels.
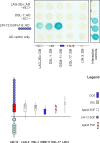
Figure 8. OSM-11 and C. elegans DSL Ligands Interact with LIN-12 Notch Extracellular Domain EGF Repeats in the Two-Hybrid System
DSL-1, OSM-11, LAG-2 extracellular domain (LAG-2Ex), EGL-17, or LIN-3 was fused to the GAL4 DNA binding domain (DB); the first six LIN-12 EGF repeats were fused to the GAL4 activation domain (AD). Pairwise interactions were tested with the yeast two-hybrid assay; positive interactions are indicated by blue staining. Both Notch DSL ligands and OSM-11 interacted with LIN-12 EGF repeats, whereas no interaction of LIN-3 EGF or EGL-17 FGF with LIN-12 Notch receptor EGF repeats was detected. LIN-12::DB fusion proteins exhibited strong self-activation (unpublished data); therefore, reciprocal fusions were not tested. Interaction controls are: (1) empty vectors; (2) DB-pRb and AD-E2F; (3) DB-Fos and AD-Jun; (4) Gal4p and pPC86; and (5) DB-DP1 and AD-E2F1.
Figure 9. Model: C. elegans DSL and DOS Proteins May Act as Ligands for Notch Receptors
Canonical Notch ligands in Drosophila contain both DSL domains and DOS motifs as do some vertebrate Notch ligands (e.g., Delta). However, classical Notch ligands from C. elegans and several vertebrate Notch ligands contain a DSL domain, but lack DOS-motif EGF repeats (e.g., LAG-2 or DLL3). The C. elegans proteins characterized in this study (e.g., OSM-11) and the two presumptive vertebrate ligands DLK1 and EGFL9/DLK2 lack DSL domains, but contain DOS motifs. In the simplest model, both a DOS motif and DSL domain are required for coordinated Notch receptor activation. These could act in cis in canonical Notch receptors like Drosophila Delta or in trans in the case of LAG-2 and OSM-11. Overexpression of a “DOS-only” or a “DSL-only” ligand may inhibit Notch receptor activation by competition with canonical ligands containing both a DSL domain and a DOS motif, such as Jagged1 or Delta. This model is consistent with osm-11(lf) animals having phenotypic defects usually associated with Notch loss of function. We do not exclude other possible scenarios; see Discussion for details.
Similar articles
- C. elegans Notch signaling regulates adult chemosensory response and larval molting quiescence.
Singh K, Chao MY, Somers GA, Komatsu H, Corkins ME, Larkins-Ford J, Tucey T, Dionne HM, Walsh MB, Beaumont EK, Hart DP, Lockery SR, Hart AC. Singh K, et al. Curr Biol. 2011 May 24;21(10):825-34. doi: 10.1016/j.cub.2011.04.010. Epub 2011 May 5. Curr Biol. 2011. PMID: 21549604 Free PMC article. - Transcriptional control of Notch signaling by a HOX and a PBX/EXD protein during vulval development in C. elegans.
Takács-Vellai K, Vellai T, Chen EB, Zhang Y, Guerry F, Stern MJ, Müller F. Takács-Vellai K, et al. Dev Biol. 2007 Feb 15;302(2):661-9. doi: 10.1016/j.ydbio.2006.09.049. Epub 2006 Oct 4. Dev Biol. 2007. PMID: 17084835 - Interchangeability of Caenorhabditis elegans DSL proteins and intrinsic signalling activity of their extracellular domains in vivo.
Fitzgerald K, Greenwald I. Fitzgerald K, et al. Development. 1995 Dec;121(12):4275-82. doi: 10.1242/dev.121.12.4275. Development. 1995. PMID: 8575327 - Vulval development.
Sternberg PW. Sternberg PW. WormBook. 2005 Jun 25:1-28. doi: 10.1895/wormbook.1.6.1. WormBook. 2005. PMID: 18050418 Free PMC article. Review. - LIN-12/Notch signaling in C. elegans.
Greenwald I. Greenwald I. WormBook. 2005 Aug 8:1-16. doi: 10.1895/wormbook.1.10.1. WormBook. 2005. PMID: 18050403 Free PMC article. Review.
Cited by
- An extracellular region of Serrate is essential for ligand-induced cis-inhibition of Notch signaling.
Fleming RJ, Hori K, Sen A, Filloramo GV, Langer JM, Obar RA, Artavanis-Tsakonas S, Maharaj-Best AC. Fleming RJ, et al. Development. 2013 May;140(9):2039-49. doi: 10.1242/dev.087916. Development. 2013. PMID: 23571220 Free PMC article. - Pleiotropic Role of Notch Signaling in Human Skin Diseases.
Gratton R, Tricarico PM, Moltrasio C, Lima Estevão de Oliveira AS, Brandão L, Marzano AV, Zupin L, Crovella S. Gratton R, et al. Int J Mol Sci. 2020 Jun 13;21(12):4214. doi: 10.3390/ijms21124214. Int J Mol Sci. 2020. PMID: 32545758 Free PMC article. Review. - Dicarbonyl/L-xylulose reductase (DCXR) producing xylitol regulates egg retention through osmolality control in Caenorhabditis elegans.
Kim YN, Kim SH, Son LT, Ahnn J, Lee SK. Kim YN, et al. Anim Cells Syst (Seoul). 2022 Oct 6;26(5):223-231. doi: 10.1080/19768354.2022.2126886. eCollection 2022. Anim Cells Syst (Seoul). 2022. PMID: 36275448 Free PMC article. - Notch signaling.
Kopan R. Kopan R. Cold Spring Harb Perspect Biol. 2012 Oct 1;4(10):a011213. doi: 10.1101/cshperspect.a011213. Cold Spring Harb Perspect Biol. 2012. PMID: 23028119 Free PMC article. Review. - Misoprostol-Induced Modification of the Notch Signaling Pathway in the Human Cervix.
Avci S, Simsek M, Soylu H, Ustunel I. Avci S, et al. Reprod Sci. 2019 Jul;26(7):909-917. doi: 10.1177/1933719118799208. Epub 2018 Oct 3. Reprod Sci. 2019. PMID: 30278829
References
- Fleming RJ, Scottgale TN, Diederich RJ, Artavanis-Tsakonas S. The gene Serrate encodes a putative EGF-like transmembrane protein essential for proper ectodermal development in Drosophila melanogaster . Genes Dev. 1990;4:2188–2201. - PubMed
- Kopczynski CC, Alton AK, Fechtel K, Kooh PJ, Muskavitch MA. Delta, a Drosophila neurogenic gene, is transcriptionally complex and encodes a protein related to blood coagulation factors and epidermal growth factor of vertebrates. Genes Dev. 1988;2:1723–1735. - PubMed
- Thomas U, Speicher SA, Knust E. The Drosophila gene Serrate encodes an EGF-like transmembrane protein with a complex expression pattern in embryos and wing discs. Development. 1991;111:749–761. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials