Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila - PubMed (original) (raw)
Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila
Yuzuru Imai et al. EMBO J. 2008.
Abstract
Dominant mutations in leucine-rich repeat kinase 2 (LRRK2) are the most frequent molecular lesions so far found in Parkinson's disease (PD), an age-dependent neurodegenerative disorder affecting dopaminergic (DA) neuron. The molecular mechanisms by which mutations in LRRK2 cause DA degeneration in PD are not understood. Here, we show that both human LRRK2 and the Drosophila orthologue of LRRK2 phosphorylate eukaryotic initiation factor 4E (eIF4E)-binding protein (4E-BP), a negative regulator of eIF4E-mediated protein translation and a key mediator of various stress responses. Although modulation of the eIF4E/4E-BP pathway by LRRK2 stimulates eIF4E-mediated protein translation both in vivo and in vitro, it attenuates resistance to oxidative stress and survival of DA neuron in Drosophila. Our results suggest that chronic inactivation of 4E-BP by LRRK2 with pathogenic mutations deregulates protein translation, eventually resulting in age-dependent loss of DA neurons.
Figures
Figure 1
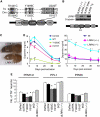
dLRRK regulates function and maintenance of DA neuron. (A) A schematic of dLRRK and hLRRK2 domain structures. (B) Western blot analysis showing loss of dLRRK protein expression in dLRRK(−/−). Brain tissues of 3-day-old adult flies were analysed using anti-dLRRK antibody, which recognizes the N-terminal part of dLRRK. Diagram indicates the location of P-element insertion. (C) A phenotype of malformed abdomen observed with incomplete penetrance in dLRRK(−/−) females. dLRRK(+/−) female shows a normal phenotype. (D) Fly heads of dLRRK Tg driven by Ddc-Gal4 (left) or dLRRK mutant animals (right) were used to prepare tissue extracts for dopamine measurement. Ddc-Gal4/+ and w − serves as controls for Tg and dLRRK mutant, respectively. The values represent means±s.e. from five male fly heads in three independent measurements (Asterisk in left and right panels, P<0.01 and _P_<0.001, respectively). (**E**) Quantification of TH+ DA neuron number in the PPM 1 and 2, PPL1 and PPM3 clusters in 60-day-old males of the indicated Tg animals driven by _TH-Gal4_. PPM1 and PPM2 cluster neurons were counted together. Data were shown as means±s.d. (*_P_<0.01 versus _TH-Gal4_>DsRed control, _n_=12 for dLRRK(−/−); _n_=10 for the others).
Figure 2
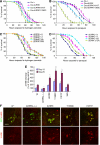
dLRRK regulates stress resistance. (A) Response of dLRRK Tg flies to H2O2 treatment. Error bars show s.d. from four repeated experiments. *P<0.05, Y1383C and I1915T versus _Da-Gal4_/+ control (all, _n_=60). (**B**) Response of dLRRK Tg to paraquat treatment. Error bars show s.d. from three repeated experiments. *_P_<0.05, Y1383C (_n_=48) and I1915T (_n_=48) versus _Da-Gal4_/+ (_n_=84). (**C**) Response of _dLRRK_(−/−) (_n_=85), _dLRRK_(_Df_/−) (_n_=84) and _dLRRK RNAi_ flies (_n_=85) to H2O2 treatment. Error bars show s.d. from three repeated experiments. _P_<0.01 versus _dLRRK_(+/+) control (_n_=89) for all values at 61–128 h. _Df_/+ and _dLRRK_(+/+) serve as controls. Transgene and RNAi expressions were directed by _Da-Gal4_ in A–C. (**D**) Response of _dLRRK_(−/−) (_n_=48) and _dLRRK_(_Df_/−) flies (_n_=46) to paraquat treatment. Error bars show s.d. from three repeated experiments. _P_<0.001, versus _dLRRK_(+/+) (_n_=72) for all values at 24–73 h. (**E**) Statistical analysis of 4-HNE levels among the indicated genotypes crossed with a _TH-Gal4_>UAS-GFP line at 10- and 20-day of age raised at 29°C. 4-HNE levels from 25–30 TH+ neurons in the PPL1 clusters were quantified after normalization with GFP signal. *P<0.05; **_P_<0.01; ***_P_<0.001 versus _w−_ × _TH-Gal4_>GFP cross (_w_−). (F) Representative images of PPL1 clusters of the indicated genotypes double-stained for GFP (green) and 4-HNE (red). Scale bar=20 μm.
Figure 3
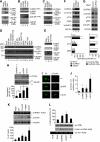
dLRRK and hLRRK2 phosphorylate 4E-BP. (A–C) In vitro kinase assays using dLRRK and d4E-BP (A, B) or hLRRK2 and h4E-BP1 (C) as kinase–substrate pairs. Mock immunoprecipitate (IP) serves as control. Autoradiography (P32), western blot (WB) and Coomassie brilliant blue (CBB) staining of the gels are shown. The asterisk in C marks a putative truncated form of hLRRK2 often observed in the IP fraction. (D, E) In vitro kinase assay using hLRRK2 (D) or dLRRK (E) as the kinase and a series of wild-type (WT) and mutant h4E-BP1 (D) or d4E-BP (E) as substrates. Mock IP serves as kinase control. CBB: protein loading control. The Ser or Thr residues mutated to Ala are indicated. (F, G) Western blot analysis showing effects of altered hLRRK2 activities on endogenous h4E-BP1 phosphorylation in 293T cells, which were starved for 24 h and then stimulated with 1 μg/ml insulin for 30 min. (F) Overexpression of WT and I2020T mutant hLRRK2 increased h4E-BP1 phosphorylation at T37/T46 and T70. Mock transfection serves as control. (G) Knockdown of hLRRK2 by RNAi reduced h4E-BP1 phosphorylation at T37/T46 and T70. Control: a non-targeting siRNA. Graphs show relative levels of p-T37/T46, p-T70 and p-S65 after normalization with total h4E-BP1 level. Values represent means±s.d. from three experiments (*P<0.05; **P<0.01 in Bonferroni/Dunn test). (H) dLRRK influences d4E-BP phosphorylation in vivo. d4E-BP protein was immunoprecipitated with a d4E-BP antibody from fly brain extracts of the indicated genotypes. Immunoprecipitated d4E-BP was detected by western blot with p-T37/T46 (upper) and total d4E-BP (lower) antibodies. Bands corresponding to phosphorylated (β) and non-phosphorylated forms (α) of d4E-BP are indicated in the total d4E-BP western. Graph shows relative level of p-T37/T46 after normalization with total d4E-BP level. Values represent means±s.d. from three experiments (*P<0.05; **P<0.01 in Bonferroni/Dunn test). (I, J) Immunohistochemical analysis showing that dLRRK promotes d4E-BP phosphorylation. Adult brain TH-positive neurons were co-stained with anti-TH and anti-p-T37/T46 in control _w_− or dLRRK Tg crossed with TH-Gal4. Representative images are shown in I. The p-T37/T46 signals were quantified after normalization with TH signals. Values represent means±s.d. from three independent experiments (*P<0.05; **P<0.01). (K) Western blot analysis showing effects of pathogenic hLRRK2 mutations on h4E-BP1 phosphorylation at T37/T46 sites in serum-starved 293T cells. The graph shows quantification of the relative level of p-T37/T46 after normalization with total h4E-BP1 level. Values represent means±s.d. from three independent experiments (*P<0.05; **P<0.01 versus hLRRK2 WT). (L) hLRRK2 stimulates protein synthesis in vitro in a kinase activity-dependent manner. Immunopurified FLAG–hLRRK2 WT, I2020T and 3KD proteins together with capped firefly luciferase mRNA were incubated in rabbit reticulocyte lysate (Promega) for 2.5 h at 30 °C. The activity of luciferase translated in the lysate was measured (graph, means±s.d. from three independent experiments). *P<0.05 versus mock. Western blot and RT–PCR was performed for estimation of protein and mRNA levels in the lysate. PCR without RT (PCR) serves as a negative control for RT–PCR.
Figure 4
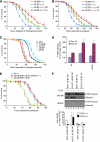
The eIF4E/4E-BP pathway is important for handling oxidative stress. (A, B) d4E-BP TA confers H2O2 (A) and paraquat (B) resistance. d4E-BP WT or TA mutant were expressed in the d4E-BP(−/−) background under DA-Gal4 control. *P<0.05, _d4E-BP_(−/−); _TA_ (_n_=76 in A; _n_=78 in B) versus _d4E-BP_(−/−); _WT_ (_n_=80 in A; _n_=82 in B). (**C**) Oxidative stress responses of dLRRK, deIF4E and d4E-BP Tg flies driven by _Da-Gal4_. deIF4E Tg (_n_=72) and the dLRRK Y1383C (_n_=72) or I1915T (_n_=85) Tg were more sensitive to H2O2 treatment than the GFP Tg control (_n_=90) (_P_<0.01 for all values at 72–120 h), whereas d4E-BP Tg (_n_=90) was more resistant than the control (_P_<0.05 versus GFP Tg for the values at 72–144 h). (**D**) Overexpression of deIF4E or dLRRK increases 4-HNE levels in DA neurons. _TH-Gal4_>GFP was crossed with the indicated genotypes as shown in Figure 2E. Values represent means±s.d. (*P<0.05; **_P_<0.01 versus age-matched _w_−, _n_=25–30). Error bars in (A–D) show s.d. from three repeated experiments. (**E**) Reduction of deIF4E function suppresses vulnerability of dLRRK I1915T to oxidative stress induced by paraquat. dLRRK I1915T was expressed in the _deIF4E_(+/+) or _deIF4E_(+/−) background with _DA-Gal4_. *_P_<0.006, _Da_>dLRRK I1915T/deIF4E (+/−) (_n_=74) versus _Da_>dLRRK I1915T (_n_=75). (F) m7GTP pull-down assay showing the effect of dLRRK I1915T on free eIF4E levels. eIF4E was precipitated using m7GTP-sepharose from the brain tissues of flies with or without 5% H2O2 treatment for 24 h. eIF4E-bound (m7GTP-bound) and free (unbound) 4E-BP were estimated. Graph shows the percentage of free 4E-BP in total 4E-BP after normalization with m7GTP-bound eIF4E level. Values represent means±s.d. from three experiments (*P<0.05; **P<0.01 versus GFP; 4E-BP WT in corresponding treatment).
Figure 5
4E-BP overexpression suppresses dysfunction and degeneration of DA neuron induced by mutant dLRRK. (A) d4E-BP co-expression rescues dLRRK overexpression-induced DA neuron loss (*P<0.01 in Student's _t_-test, _n_=12). _TH-Gal4_ was used to direct transgene expression and 60-day-old flies aged at 25 °C were analysed. (**B**) d4E-BP TA protects against DA neuron loss in dLRRK I1915T Tg flies. _TH-Gal4_ was used to direct transgene expression and 30-day-old flies aged at 29 °C were analysed. *_P_<0.05, _n_=14. (**C**) _dLRRK_ LOF rescues deIF4E overexpression-induced DA neuron loss (*_P_<0.01, _n_=12). _TH-Gal4_ crosses were analysed as in (A). (**D**) Pan-neuronal expression of dLRRK I1915T under _elav-Gal4_ control leads to gradual motor defect with age. The loss of climbing ability in dLRRK I1915T-expressing flies was rescued by 4E-BP TA expression. GFP serves as control. The values represent means±s.d. from 20 trials (_n_=30; *_P_<0.05 versus GFP; #_P_<0.01 versus I1915T by one-way ANOVA with Bonferroni/Dunn test). The genotypes are as follows: _elav-Gal4_>UAS-GFP (GFP), _elav-Gal4_>UAS-4E-BP WT (4E-BP WT), _elav-Gal4_>UAS-4E-BP TA (4E-BP TA), _elav-Gal4_>UAS-dLRRK I1915T (I1915T), _elav-Gal4_>UAS-dLRRK I1915T/UAS-4E-BP WT (I1915T; 4E-BP WT), _elav-Gal4_>UAS-dLRRK I1915T/UAS-4E-BP TA (I1915T; 4E-BP TA). Male flies were used for the assay.
Figure 6
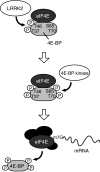
A model depicting 4E-BP phosphorylation by dLRRK. Phosphorylation of 4E-BP at T37/T46 residues by LRRK2 (upper) facilitates subsequent phosphorylation at T70 and S65 (middle). Hyperphosphorylated 4E-BP is released from mRNA cap-binding protein eIF4E, which leads to the formation of an initiation factor complex including eIF4E for protein translation (lower).
Similar articles
- Phosphorylation of 4E-BP1 in the mammalian brain is not altered by LRRK2 expression or pathogenic mutations.
Trancikova A, Mamais A, Webber PJ, Stafa K, Tsika E, Glauser L, West AB, Bandopadhyay R, Moore DJ. Trancikova A, et al. PLoS One. 2012;7(10):e47784. doi: 10.1371/journal.pone.0047784. Epub 2012 Oct 17. PLoS One. 2012. PMID: 23082216 Free PMC article. - The Parkinson's disease associated LRRK2 exhibits weaker in vitro phosphorylation of 4E-BP compared to autophosphorylation.
Kumar A, Greggio E, Beilina A, Kaganovich A, Chan D, Taymans JM, Wolozin B, Cookson MR. Kumar A, et al. PLoS One. 2010 Jan 15;5(1):e8730. doi: 10.1371/journal.pone.0008730. PLoS One. 2010. PMID: 20090955 Free PMC article. - LRRK2 kinase regulates synaptic morphology through distinct substrates at the presynaptic and postsynaptic compartments of the Drosophila neuromuscular junction.
Lee S, Liu HP, Lin WY, Guo H, Lu B. Lee S, et al. J Neurosci. 2010 Dec 15;30(50):16959-69. doi: 10.1523/JNEUROSCI.1807-10.2010. J Neurosci. 2010. PMID: 21159966 Free PMC article. - The synaptic function of LRRK2.
Lee S, Imai Y, Gehrke S, Liu S, Lu B. Lee S, et al. Biochem Soc Trans. 2012 Oct;40(5):1047-51. doi: 10.1042/BST20120113. Biochem Soc Trans. 2012. PMID: 22988863 Review. - Phosphorylation of the cap-binding protein eIF4E by the MAPK-activated protein kinase Mnk1.
Pyronnet S. Pyronnet S. Biochem Pharmacol. 2000 Oct 15;60(8):1237-43. doi: 10.1016/s0006-2952(00)00429-9. Biochem Pharmacol. 2000. PMID: 11007962 Review.
Cited by
- Defects in mRNA Translation in LRRK2-Mutant hiPSC-Derived Dopaminergic Neurons Lead to Dysregulated Calcium Homeostasis.
Kim JW, Yin X, Jhaldiyal A, Khan MR, Martin I, Xie Z, Perez-Rosello T, Kumar M, Abalde-Atristain L, Xu J, Chen L, Eacker SM, Surmeier DJ, Ingolia NT, Dawson TM, Dawson VL. Kim JW, et al. Cell Stem Cell. 2020 Oct 1;27(4):633-645.e7. doi: 10.1016/j.stem.2020.08.002. Epub 2020 Aug 25. Cell Stem Cell. 2020. PMID: 32846140 Free PMC article. - Light-driven activation of mitochondrial proton-motive force improves motor behaviors in a Drosophila model of Parkinson's disease.
Imai Y, Inoshita T, Meng H, Shiba-Fukushima K, Hara KY, Sawamura N, Hattori N. Imai Y, et al. Commun Biol. 2019 Nov 22;2:424. doi: 10.1038/s42003-019-0674-1. eCollection 2019. Commun Biol. 2019. PMID: 31799427 Free PMC article. - Synaptic dysfunction in genetic models of Parkinson's disease: a role for autophagy?
Plowey ED, Chu CT. Plowey ED, et al. Neurobiol Dis. 2011 Jul;43(1):60-7. doi: 10.1016/j.nbd.2010.10.011. Epub 2010 Oct 20. Neurobiol Dis. 2011. PMID: 20969957 Free PMC article. Review. - LRRK2: cause, risk, and mechanism.
Paisán-Ruiz C, Lewis PA, Singleton AB. Paisán-Ruiz C, et al. J Parkinsons Dis. 2013;3(2):85-103. doi: 10.3233/JPD-130192. J Parkinsons Dis. 2013. PMID: 23938341 Free PMC article. Review. - Senolytic and senomorphic secondary metabolites as therapeutic agents in Drosophila melanogaster models of Parkinson's disease.
Miller SJ, Darji RY, Walaieh S, Lewis JA, Logan R. Miller SJ, et al. Front Neurol. 2023 Sep 28;14:1271941. doi: 10.3389/fneur.2023.1271941. eCollection 2023. Front Neurol. 2023. PMID: 37840914 Free PMC article. Review.
References
- Fadden P, Haystead TA, Lawrence JC Jr (1997) Identification of phosphorylation sites in the translational regulator, PHAS-I, that are controlled by insulin and rapamycin in rat adipocytes. J Biol Chem 272: 10240–10247 - PubMed
- Feany MB, Bender WW (2000) A Drosophila model of Parkinson's disease. Nature 404: 394–398 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR054926/AR/NIAMS NIH HHS/United States
- R21 NS056878/NS/NINDS NIH HHS/United States
- R01 AR054926-01/AR/NIAMS NIH HHS/United States
- R21 NS056878-01/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous