The transcription factor PLZF directs the effector program of the NKT cell lineage - PubMed (original) (raw)
The transcription factor PLZF directs the effector program of the NKT cell lineage
Adam K Savage et al. Immunity. 2008.
Abstract
The transcriptional control of CD1d-restricted NKT cell development has remained elusive. We report that PLZF (promyelocytic leukemia zinc finger, Zbtb16), a member of the BTB/POZ-ZF family of transcription factors that includes the CD4-lineage-specific c-Krox (Th-POK), is exquisitely specific to CD1d-restricted NKT cells and human MR1-specific MAIT cells. PLZF was induced immediately after positive selection of NKT cell precursors, and PLZF-deficient NKT cells failed to undergo the intrathymic expansion and effector differentiation that characterize their lineage. Instead, they preserved a naive phenotype and were directed to lymph nodes. Conversely, transgenic expression of PLZF induced CD4(+) thymocytes to acquire effector differentiation and migrate to nonlymphoid tissues. We suggest that PLZF is a transcriptional signature of NKT cells that directs their innate-like effector differentiation during thymic development.
Figures
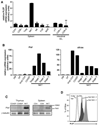
Figure 1. PLZF Expression Is Specific to NKT Cells
(A) Purified subsets of splenocytes and intestinal intraepithelial lymphocytes as indicated were assessed for PLZF mRNA expression by qRT-PCR. Data shown as mean +/− SD or triplicate PCR values. (B) PLZF and c-Krox mRNA expression in thymic subsets and thymic NKT developmental stages as indicated. DN, CD4−8− double negative; DP, CD4+8+ double positive; trnsl CD4, transitional CD4+8lowCD24highTCRβhigh; CD4SP, CD4+8− single positive; NKT stage 0, CD24highCD69high; NKT stage 1, CD24lowCD44lowNK1.1−; NKT stage 2, CD24lowCD44highNK1.1−; NKT stage 3, CD24lowCD44highNK1.1+. Data shown as mean +/− SD of triplicate PCR analysis and representative of 2–3 independent experiments. (C) PLZF western blot analysis of sorted CD1d-αGalCer+ NKT cells and conventional CD4 and CD8 cells (1x106 cells per lane) in the thymus and spleen, as indicated. Anti γ-tubulin was used as a loading control. Data are representative of 3 experiments (D) Intracellular flow cytometry with anti-PLZF mAb D-9 of thymic NKT stage 1 + 2 cells (Tetr+NK1.1−, grey shaded), stage 3 (Tetr+NK1.1+; black shaded), and conventional T cells (TCRb+Tetr−), as indicated. The isotype control is the dashed histogram. Data are representative of 4 experiments.
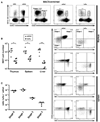
Figure 2. Luxoid Mice Have a Selective Defect in NKT Cell Development
(A) The frequency of NKT cells in the thymus, spleen, and liver of Luxoid (_Zbtb_16lu/lu) mice compared to their wt/wt littermate controls. NKT cells were gated (as shown) as CD24lowCD1d-αGC+ in the thymus and CD1d-αGC+TCRβ+ (or CD1d-αGC+B220− in some experiments) in the spleen and liver. Data are representative of 6 pairs of mice. (B) Summary of NKT cell absolute numbers in individual thymuses, spleens, and livers of wt/wt and lu/lu littermates. * p<=0.05, ** p<=0.01. (C) Unconventional intestinal IELs in Luxoid mice and their wt littermates. Upper row, frequency of αβ and γδ T cells among CD3+ gated IELs; lower row, frequency of CD8αα and CD8αβ T cells among TCRβ+ gated IELs. Absolute numbers of IELs were similar in wt and lu/lu mice. Data are representative of 5 littermate pairs in two independent experiments.
Figure 3. Mixed Bone Marrow Chimeras Reveal a Cell-intrinsic NKT Cell Defect
(A) CD45.2 lu/lu and CD45.1 wt/wt bone marrows were mixed at a 1:1 ratio and transferred into lethally irradiated Jα18−/− hosts. 6–8 weeks later, reconstituted animals were analyzed by flow cytometry. Individual thymus and spleen from the chimeras were MACS-enriched using CD1d-αGC tetramers prior to FACS analysis of NKT developmental subsets. Upper row, leftmost panel shows the relative proportion of CD45.2 (lu/lu) and CD45.1 (wt) NKT lineage cells in the thymus; right panels show the proportion of CD24high (stage 0) and CD24low (stage1, 2 and 3) cells in the total population (2nd panel from left) and in gated CD45.1 (wt) (3rd panel) and CD45.2 (lu/lu) (4th panel) CD1d-αGC+ cells. Wt and lu/lu CD24low NKT cells are further analyzed based on CD44 and NK1.1 expression, or CD44 and CD4 expression, as indicated for the thymus and spleen (bottom 2–5 rows). Data are representative of 6 individual chimeras. (B) Summary of CD1d-αGC+ cell numbers in the wt and lu/lu compartments of individual mixed chimeras. Data are normalized based on the relative level of reconstitution by CD45.1 and CD45.2 bone marrows (e.g. NKT cell number in the wt CD45.1 compartment is divided by the CD45.1/(CD45.1+CD45.2) ratio measured for total lymphocytes. On average, the wt and lu/lu bone marrows contributed equally to the total lymphocyte population. (C) Summary of the ratios of wt/wt over lu/lu CD1d-αGC+ cells at different stages of thymic development. Data are normalized based on the relative level of CD45.1 and CD45.2 reconstitution.
Figure 4. PLZF-deficient NKT Cells Revert to a Naïve Phenotype
(A) Summary of the percentages of wt/wt and lu/lu cells among CD1d-αGalCer+ cells in various tissues of individual mixed bone marrow chimeras. The percentages are normalized relative to the level of reconstitution of the total lymphocyte population by wt/wt and lu/lu cells, as assessed by CD45.1 and CD45.2 staining. For example, the ratio of CD45.1 CD1d-αGalCer+ cells over the total CD45.1 lymphocyte population is divided by (ratio of CD45.1+CD1d-αGalCer+ cells over the total CD45.1 lymphocyte population + ratio of CD45.2+CD1d-αGalCer+ cells over the total CD45.2 lymphocyte population) and multiplied by 100. *** p<=0.0001. (B) Expression of CD62L by wt/wt and lu/lu CD1d-αGalCer+ cells in various tissues of mixed bone marrow chimeras. Data are representative of 5 chimeras. (C) Cytokine secretion by Luxoid NKT cells. Left, cytokines released after stimulation of 5000 CD1d-αGalCer tetramer-sorted splenocytes (pooled from luxoid or wt littermates) with anti-CD3 + human IL-2; grey bars: lu/lu cells, black bars: +/+ littermate cells. A major increase in the IL-2/IL-4 ratio was also observed in a separate experiment comparing tetramer-sorted lu/lu vs wt splenocytes stimulated with αGalCer-loaded DC. Right, stimulation of 3x105 CD19- and CD8-depleted Vα14-Jα18 transgenic ‘414’ splenocytes with αGalCer-pulsed bone-marrow derived dendritic cells; grey bars: lu/lu 414 cells, black bars: +/+ 414 littermate cells. Supernatants were recovered after 2 days and values are expressed in pg/ml. Data are from an individual luxoid/littermate pair.
Figure 5. Transgenic Expression of PLZF converts CD4 thymocytes into effector cells
(A) Left, CD4/CD8 FACS dot plots of lymphocytes from various organs of CD4 promoter-PLZF transgenic and wild type littermates (CD1d-αGC Tetramer-positive cells are gated out). Right, CD44/CD62L expression by CD4 cells of indicated organs gated as shown on the left panels. Data representative of two independent transgenic lines 1797 and 1960. (B) PLZF transgenic CD4 cells in competitive chimeras. Wild-type (CD45.1+) and PLZF Tg (CD45.2+, line 1797) bone marrow cells were mixed 1:1 to reconstitute lethally irradiated wild type recipients. Thymus and spleen cells were stained as indicated (left panels) to gate CD4 and CD8 T cells for analysis of CD44 and CD45.1 (middle and right panels) Chimeras were analyzed 6.5 weeks after transfer. Data are representative of four individual chimeras. (C) Cell cycle analysis 3 hours after in vivo injection of 100µg of 5-ethynyl-2'-deoxyuridine (EdU) into mixed bone marrow chimeras. Edu incorporation was detected by intracellular flow cytometry for DP (left panels) and CD4 single positive (right panels) thymocytes of wt (top panels) and PLZF-Tg (bottom panels) origin, as indicated. Similar values were obtained for splenic CD4 cells (not shown). Data are representative of three individual chimeras and two independent experiments. (D) Frequency and tissue distribution of NKT cells in PLZF-transgenic mice. Data are from the same mixed bone marrow chimeras as in Fig. 5B and represent percentages of NKT cells among wild-type (CD45.1+) and PLZF Tg (CD45.2+) lymphocytes.
Figure 6. Cytokine profile of PLZF Transgenic CD4 T cells
(A) Intracellular flow cytometry with anti-PLZF mAb D-9 of thymic PLZF-transgenic (line 1797) CD4 SP cells compared with wt CD4 SP and with wt NKT stage 1 + 2 cells thymocytes (Tetr+NK1.1−), as indicated. (B) Decreased IL-2 secretion by PLZF-transgenic CD4 T cells. CD4 cells were MACS-enriched from spleens of PLZF-transgenic and littermate and stimulated with anti-CD3 + human IL-2 for two days. Values in pg/ml for 2 pairs of wt and Tg littermates, as indicated. (C) Intracellular flow cytometry for IL-4 and IFN-γ in littermate and PLZF transgenic mice. Splenocytes were prepared and stimulated as in A, with the addition of brefeldin A for the last six hours of stimulation. Three pairs of wt and Tg littermates were analyzed. The percentages of dual IL-4/ IFN-γ producers were 24.5, 17.3, 26.1 in transgenics vs 3.97, 4.92, 7.34 in wt.
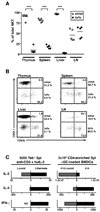
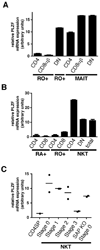
Figure 7. PLZF Expression in Human NKT and MAIT Cells and in SAP-deficient NKT thymocytes
(A) Vα7.2+CD45RO+CD161+ MAIT cells were purified from fresh human PBL according to their expression of CD4 or CD8β. The DN subset of MAIT cells is defined as CD4− CD8β−. Control Vα7.2−RO+ memory cells were purified according to expression of CD4 and CD8β. The Vα7.2−RO+CD4−CD8β− (DN RO+) population is enriched in CD1d-restricted unconventional T cells. (B) NKT cells were MACS-enriched from fresh human PBL using CD1d-αGC tetramer and FACS-sorted into total NKT cells or their CD4 and DN subsets. The NKT-depleted MACS fraction was used to FACS purify naïve (RA+) and memory (RO+) CD4 and CD8 conventional T cells. (C) PLZF expression by SAP-deficient ‘stage 0’ NKT thymocytes. 2000 CD1d-αGC+thymocytes were sorted at different stages of NKT development for wild-type mice and at the arrested ‘stage 0’ for SAP-deficient mice. PLZF mRNA expression was measured by quantitative qRT-PCR. Data summarize two independent experiments. Control CD4 single positive (CDSP) are shown as negative control.
Similar articles
- Innate PLZF+CD4+ αβ T cells develop and expand in the absence of Itk.
Prince AL, Watkin LB, Yin CC, Selin LK, Kang J, Schwartzberg PL, Berg LJ. Prince AL, et al. J Immunol. 2014 Jul 15;193(2):673-87. doi: 10.4049/jimmunol.1302058. Epub 2014 Jun 13. J Immunol. 2014. PMID: 24928994 Free PMC article. - Promyelocytic leukemia zinc finger turns on the effector T cell program without requirement for agonist TCR signaling.
Savage AK, Constantinides MG, Bendelac A. Savage AK, et al. J Immunol. 2011 May 15;186(10):5801-6. doi: 10.4049/jimmunol.1100119. Epub 2011 Apr 8. J Immunol. 2011. PMID: 21478405 Free PMC article. - TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity.
Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, Pandolfi PP, Bendelac A, von Boehmer H. Kreslavsky T, et al. Proc Natl Acad Sci U S A. 2009 Jul 28;106(30):12453-8. doi: 10.1073/pnas.0903895106. Epub 2009 Jul 15. Proc Natl Acad Sci U S A. 2009. PMID: 19617548 Free PMC article. - Development of PLZF-expressing innate T cells.
Alonzo ES, Sant'Angelo DB. Alonzo ES, et al. Curr Opin Immunol. 2011 Apr;23(2):220-7. doi: 10.1016/j.coi.2010.12.016. Epub 2011 Jan 21. Curr Opin Immunol. 2011. PMID: 21257299 Free PMC article. Review. - CD4 CTL: living up to the challenge.
Cheroutre H, Husain MM. Cheroutre H, et al. Semin Immunol. 2013 Nov 15;25(4):273-81. doi: 10.1016/j.smim.2013.10.022. Epub 2013 Nov 15. Semin Immunol. 2013. PMID: 24246226 Free PMC article. Review.
Cited by
- The origin and role of innate lymphoid cells in the lung.
Lai DM, Shu Q, Fan J. Lai DM, et al. Mil Med Res. 2016 Aug 19;3:25. doi: 10.1186/s40779-016-0093-2. eCollection 2016. Mil Med Res. 2016. PMID: 27547445 Free PMC article. Review. - Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions.
Brennan PJ, Brigl M, Brenner MB. Brennan PJ, et al. Nat Rev Immunol. 2013 Feb;13(2):101-17. doi: 10.1038/nri3369. Epub 2013 Jan 21. Nat Rev Immunol. 2013. PMID: 23334244 Review. - BCL11B is positioned upstream of PLZF and RORγt to control thymic development of mucosal-associated invariant T cells and MAIT17 program.
Drashansky TT, Helm EY, Curkovic N, Cooper J, Cheng P, Chen X, Gautam N, Meng L, Kwiatkowski AJ, Collins WO, Keselowsky BG, Sant'Angelo D, Huo Z, Zhang W, Zhou L, Avram D. Drashansky TT, et al. iScience. 2021 Mar 13;24(4):102307. doi: 10.1016/j.isci.2021.102307. eCollection 2021 Apr 23. iScience. 2021. PMID: 33870128 Free PMC article. - PLZF Controls the Expression of a Limited Number of Genes Essential for NKT Cell Function.
Gleimer M, von Boehmer H, Kreslavsky T. Gleimer M, et al. Front Immunol. 2012 Dec 21;3:374. doi: 10.3389/fimmu.2012.00374. eCollection 2012. Front Immunol. 2012. PMID: 23267359 Free PMC article. - Inhibitory Potential of Shen-Shuai-Ling Formulation on Renal Interstitial Fibrosis via Upregulation of PLZF.
Song N, Tu H, Li Y, Xiong W, Zhang L, Liu H, Ding W, Long M, Ren D, Zhong J. Song N, et al. Evid Based Complement Alternat Med. 2022 Mar 31;2022:5967804. doi: 10.1155/2022/5967804. eCollection 2022. Evid Based Complement Alternat Med. 2022. PMID: 35399631 Free PMC article.
References
- Bendelac A, Bonneville M, Kearney JF. Autoreactivity by design: innate B and T lymphocytes. Nat Rev Immunol. 2001;1:177–186. - PubMed
- Bendelac A, Killeen N, Littman D, Schwartz RH. A subset of CD4+ thymocytes selected by MHC class I molecules. Science. 1994;263:1774–1778. - PubMed
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHMI/Howard Hughes Medical Institute/United States
- P30 DK042086/DK/NIDDK NIH HHS/United States
- R01 AI038339/AI/NIAID NIH HHS/United States
- P30 DK42086/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous