Different regulation of wild-type and mutant Cu,Zn superoxide dismutase localization in mammalian mitochondria - PubMed (original) (raw)
. 2008 Nov 1;17(21):3303-17.
doi: 10.1093/hmg/ddn226. Epub 2008 Aug 13.
Affiliations
- PMID: 18703498
- PMCID: PMC2566526
- DOI: 10.1093/hmg/ddn226
Different regulation of wild-type and mutant Cu,Zn superoxide dismutase localization in mammalian mitochondria
Hibiki Kawamata et al. Hum Mol Genet. 2008.
Abstract
The antioxidant enzyme Cu,Zn superoxide dismutase (SOD1) is predominantly localized in the cytosol, but it is also found in mitochondria. Studies in yeast suggest that apoSOD1 is imported into mitochondria and trapped inside by folding and maturation, which is facilitated by its copper chaperone for SOD1 (CCS). Here, we show that in mammalian cells, SOD1 mitochondrial localization is dictated by its folding state, which is modulated by several interconnected factors. First, the intracellular distribution of CCS determines SOD1 partitioning in cytosol and mitochondria: CCS localization in the cytosol prevents SOD1 mitochondrial import, whereas CCS in mitochondria increases it. Second, the Mia40/Erv1 pathway for import of small intermembrane space proteins participates in CCS mitochondrial import in a respiratory chain-dependent manner. Third, CCS mitochondrial import is regulated by oxygen concentration: high (20%) oxygen prevents import, whereas physiological (6%) oxygen promotes it. Therefore, SOD1 localization responds to changes in environmental conditions following redistribution of CCS, which operates as an oxygen sensor. Fourth, all of the cysteine residues in human SOD1 are critical for its retention in mitochondria due to their involvement in intramolecular disulfide bonds and in the interaction with CCS. Mutations in SOD1 are associated with autosomal dominant familial amyotrophic lateral sclerosis. Like the wild-type protein, mutant SOD1 localizes to mitochondria, where it induces bioenergetic defects. We find that the physiological regulation of mitochondrial localization is either inefficient or absent in SOD1 pathogenic mutants. We propose misfolding and aggregation of these mutants that trap them inside mitochondria.
Figures
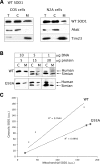
Figure 1.
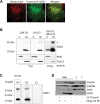
WT SOD1 localization in mammalian mitochondria. (A) Western blot of total lysate (T), cytosolic (C) and mitochondrial (M) fractions from COS or N2A neuroblastoma cells transfected with WT SOD1. Equal amounts of proteins (5 µg) were loaded in each lane, and SOD1 was detected by the SOD1 polyclonal antibody. A proportion of the WT SOD1 localizes to mitochondria in both cell types. The same blots were probed with AktK and Tim23 to exclude cytosolic protein contamination and to estimate mitochondrial loading, respectively. (B) COS cells were transfected with WT or G93A mutant SOD1 using different amounts of plasmid DNA (1, 5 and 10 µg). Proteins were loaded for the cytosolic and mitochondrial fractions as indicated (5, 15 and 30 µg), and SOD1 was detected with the SOD1 polyclonal antibody. Human SOD1 migrates higher in the gel than the endogenous simian SOD1. (C) SOD1 levels in cytosolic and mitochondrial fractions were determined by densitometry of western blot immunoreactive bands. SOD1 contents in cytosol and mitochondria were normalized to AktK and Tim23 (data not shown), respectively. The ratio between cytosolic and mitochondrial SOD1 content follows an approximately linear correlation for both WT and mutant G93A SOD1 (the _R_2 values are indicated for both WT and G93A SOD1).
Figure 2.
WT SOD1 in mitochondria is modulated by CCS localization, which depends on oxygen levels. (A) SOD1 mitochondrial localization was assessed in COS cells transfected with WT SOD1 with or without CCS-HA. CCS is detected by the anti-HA antibody in the cytosolic fraction, but not in mitochondria. SOD1 mitochondrial content is decreased by CCS co-expression by 98% when compared with cells expressing WT SOD1 alone. (B) SOD1 mitochondrial localization assessed in N2A cells co-transfected with WT SOD1 and CCS-HA. CCS is detected in the cytosol only. SOD1 is undetectable in N2A mitochondria. (C) To exclude an interference by the HA tag, COS cells were transfected with WT SOD1 and a CCS construct without the C-terminal HA. Again, using an antiserum against CCS, the protein is detected in the cytosol but not in mitochondria. Consistently, mitochondrial WT SOD1 content is reduced in COS cells co-expressing CCS when compared with cells expressing WT SOD1 alone. (D) To assess the effect of oxygen concentration on CCS localization, transfected cells were exposed either to ambient oxygen (20%) or to physiological oxygen (6%) for 48 h. Under 20% oxygen, CCS-HA is undetectable in mitochondria, but it becomes detectable in 6% oxygen. The mitochondrial content of SOD1 is increased in 6% oxygen by 60% when compared with 20% oxygen.
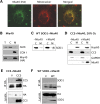
Figure 3.
Role of Mia40-Erv1 disulfide relay system in SOD1 mitochondrial import. (A) By immunocytochemistry, Mia40 (in green) co-localizes with mitochondria labeled with Mitotracker (in red), as shown by the yellow color in the merged image. (B) Mia40 expression and localization were determined in COS cells transfected with Mia40-HA. Cellular fractionation shows Mia40 localized almost exclusively in the mitochondrial fraction. Cytochrome oxidase subunit I (COXI) was used as a mitochondrial marker and AktK as a cytosolic marker. (C) SOD1 mitochondrial content is unaffected by co-expression with Mia40 when compared with empty vector control (−Mia40). (D) In cells transfected with CCS with or without Mia40 under 20% oxygen, a small proportion of CCS localizes to mitochondria only when co-expressed with Mia40. In this case, Hsp60 was used as a mitochondrial marker. (E) In cells co-expressing CCS and Mia40(HA) or empty vector, immunoprecipitation of Mia40 using the HA antibody shows that CCS co-immunoprecipitates with Mia40. (F) In cells co-expressing SOD1 and Mia40(HA) or empty vector, immunoprecipitation using the HA antibody does not show WT SOD1 co-immunoprecipitation with Mia40.
Figure 4.
Regulation of mitochondrial CCS and SOD1 content by the respiratory chain. Mitochondrial CCS and WT SOD1 contents were assessed in cells treated with the complex III inhibitor, antimycin A (AA) and compared with untreated cells (Unt). (A) AA has minimal effects on SOD1 mitochondrial content in cells transfected with WT SOD1 alone (left panel) or with Mia40 (right panel). (B) AA treatment results in approximately a 4-fold increase of mitochondrial CCS content when the protein is expressed together with Mia40 (right panel), but not when it is expressed by itself (left panel). The relative CCS mitochondrial content is shown at the bottom. (C) In cells co-expressing CCS and SOD1, AA results in an increase in the mitochondrial content of SOD1. The relative SOD1 mitochondrial content is shown at the bottom.
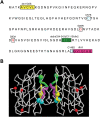
Figure 5.
Human SOD1 mutagenesis. (A) The SOD1 residues that were changed by in vitro mutagenesis to create the mutants used in the study are highlighted. Hydrophobic stretches were removed to create deletion 1 (dln1, yellow), deletion 2 (dln2, green) and deletion 3 (dln3, magenta) mutants. Four cysteine residues (C6, C57, C111, C146) were changed to serines. C6 and C111 are located within the regions encompassing dln1 and dln2, respectively. The cysteines residues forming the intramolecular disulfide bridge, C57 and C146, are indicated in blue. Pathogenic mutants G85R and G93A SOD1 are shown in red. The Frameshift SOD1 mutant has a 35 amino acid-altered C-terminus with an early truncation at amino acid 145. (B) Tridimensional representation of dimerized SOD1. The mutated residues are indicated by colors as in (A). The hydrophobic stretches of SOD1 are located at the dimer interface. C57 and C146 (in blue), which form the intramolecular disulfide bond, are faced opposing each other.
Figure 6.
Cysteine substitutions and ablation of hydrophobic stretch 2 reduced SOD1 mitochondrial localization. (A) All cysteine to serine mutations in human SOD1 result in significantly reduced mitochondrial localization when compared with WT. Asterisk indicates the cysteines involved in the intramolecular disulfide bond formation. Mouse SOD1, which naturally contains a serine at position 111, localizes to mitochondria much more abundantly than human C111S SOD1. The proportion of mitochondrial/cytosolic SOD1 is shown next to each panel. (B) Deletion of hydrophobic stretches in dln1 and dln3 mutants does not impair SOD1 mitochondrial localization, whereas the dln2 mutant is undetectable in the mitochondrial fraction. The proportion of mitochondrial/cytosolic SOD1 is shown. (C) Cytosolic fractions containing WT and dln2 SOD1 were treated with mitochondrial extracts (Ext) from untransfected COS cells. As controls, samples were incubated with ME plus protease inhibitors (Ext PI) or mitochondrial resuspension buffer alone (B). Neither WT nor dln2 SOD1 are degraded by proteases contained in the ME. (D) Detergent solubility assay to detect SOD1 aggregates in total lysates from cells expressing C6S, dln1 and WT SOD1. Only the dln1 mutant protein forms detergent-insoluble aggregates detected in the P2 fraction, whereas WT and C6S SOD1 are only found in the soluble (S1) fraction.
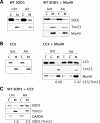
Figure 7.
Mitochondrial localization of pathogenic SOD1 mutants. (A) The mitochondrial content of WT, G93A, and G85R SOD1 was assessed with and without CCS co-expression. The co-expression of CCS reduces WT SOD1 mitochondrial localization by 96% (downward arrow), as compared to WT SOD1 expressed alone. Co-expression of CCS reduces G93A mitochondrial content by 50%, as compared to G93A SOD1 expressed alone. CCS has no effect on relative G85R SOD1 mitochondrial localization. The arrow denotes the band corresponding to human G85R SOD1, which migrates slightly above simian endogenous SOD1. (B) Cells co-transfected with G93A or G85R SOD1 and CCS, treated with (AA) or without (Unt) antimycin A. G93A SOD1 mitochondrial content is increased 300% by AA, as compared to Unt (left panels), reflecting the increase in mitochondrial CCS. G85R SOD1 is not affected by changes in CCS mitochondrial localization induced by AA (right panels). (C) Mitochondrial localization of G85R SOD1 is not affected by changes in oxygen concentration, regardless of CCS expression. (D) Mitochondria from WT, G93A and G93A/C111S SOD1-expressing cells were separated into NP40 detergent soluble and insoluble fractions. WT SOD1 is found exclusively in the detergent soluble fraction (S1), whereas the G93A mutant is largely contained in the detergent-insoluble fraction (P2). G93A/C111S mutant has a reduced detergent-insoluble component when compared with the G93A mutant. Expression of SOD1 and the levels of the mitochondrial matrix protein Hsp60 are shown by western blot of cell lysates (bottom panels). (E) Mitochondrial localization of G93A/C111S SOD1 is decreased by 20% when compared with G93A SOD1.
Figure 8.
A highly aggregatable Frameshift SOD1 mutant accumulates in mitochondria. (A) COS cells transfected with Frameshift SOD1 were immunostained with the monoclonal anti-human SOD1 antibody. This mutant (in green) co-localizes with mitochondria labeled with Mitotracker (in red), as shown by the merged image. (B) Fractionation studies of Frameshift SOD1-expressing cells, cultured in 20 or 6% oxygen, show that this mutant is undetectable in the cytosol and it accumulates exclusively in mitochondria (left panels). Upon proteasome inhibition with 20 µ
m
MG132, Frameshift SOD1 becomes detectable also in the cytosolic fraction (right panel). The amounts of protein loaded are shown at the bottom of each panel. The asterisk denotes mutant SOD1 oligomers resistant to SDS and β-mercaptoethanol. (C) In mitochondria, Frameshift SOD1 is only found in NP-40-insoluble aggregates (P2), as determined by a detergent solubility assay. (D) Intact mitochondria (Mito) from Frameshift SOD1-expressing cells were treated with (+) or without (−) proteinase K (PK). Most of the Frameshift SOD1 associated with mitochondria is still detectable after PK treatment, but is degraded when mitochondrial membranes are solubilized with Triton X, suggesting that Frameshift SOD1 is localized inside mitochondria. T, total cell lysate; C, cytosolic fraction. Hsp60 is used as mitochondrial matrix marker. GAPDH and AktK are used as cytosolic markers.
Figure 9.
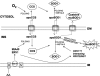
Model of regulation of SOD1 mitochondrial localization. OD1 and CCS can enter mitochondria through the general import pore of the TOM complex only in their apo-forms. The rate of folding and maturation of these proteins in the cytosol (i) determine the probability of their import in mitochondria (ii). Oxygen accelerates cytosolic maturation of CCS and consequently of SOD1, preventing mitochondrial import of both proteins. Conversely, retention in mitochondria of CCS and SOD1 is dependent upon their folding inside, which is regulated by the respiratory chain via the disulfide relay system, Mia40/Erv1, and by other unknown proteins that interact with SOD1 cysteine residues. Non-aggregatable SOD1 mutants that fail to fold due to the lack of critical cysteine residues involved in inter- or intramolecular disulfide bonds are not retained in mitochondria. On the other hand, misfolding and aggregation override the physiological regulation and determine the mitochondrial localization of pathogenic mutant SOD1. IM, mitochondrial inner membrane; IMS, intermembrane space; OM, outer membrane; AA, antimycin A; cyt c, cytochrome c.
Similar articles
- Import, maturation, and function of SOD1 and its copper chaperone CCS in the mitochondrial intermembrane space.
Kawamata H, Manfredi G. Kawamata H, et al. Antioxid Redox Signal. 2010 Nov 1;13(9):1375-84. doi: 10.1089/ars.2010.3212. Antioxid Redox Signal. 2010. PMID: 20367259 Free PMC article. Review. - Mia40 and MINOS act in parallel with Ccs1 in the biogenesis of mitochondrial Sod1.
Varabyova A, Topf U, Kwiatkowska P, Wrobel L, Kaus-Drobek M, Chacinska A. Varabyova A, et al. FEBS J. 2013 Oct;280(20):4943-59. doi: 10.1111/febs.12409. Epub 2013 Jul 22. FEBS J. 2013. PMID: 23802566 - Mia40-dependent oxidation of cysteines in domain I of Ccs1 controls its distribution between mitochondria and the cytosol.
Klöppel C, Suzuki Y, Kojer K, Petrungaro C, Longen S, Fiedler S, Keller S, Riemer J. Klöppel C, et al. Mol Biol Cell. 2011 Oct;22(20):3749-57. doi: 10.1091/mbc.E11-04-0293. Epub 2011 Aug 24. Mol Biol Cell. 2011. PMID: 21865594 Free PMC article. - The disulfide relay system of mitochondria is required for the biogenesis of mitochondrial Ccs1 and Sod1.
Reddehase S, Grumbt B, Neupert W, Hell K. Reddehase S, et al. J Mol Biol. 2009 Jan 16;385(2):331-8. doi: 10.1016/j.jmb.2008.10.088. Epub 2008 Nov 7. J Mol Biol. 2009. PMID: 19010334 - Immature copper-zinc superoxide dismutase and familial amyotrophic lateral sclerosis.
Seetharaman SV, Prudencio M, Karch C, Holloway SP, Borchelt DR, Hart PJ. Seetharaman SV, et al. Exp Biol Med (Maywood). 2009 Oct;234(10):1140-54. doi: 10.3181/0903-MR-104. Epub 2009 Jul 13. Exp Biol Med (Maywood). 2009. PMID: 19596823 Free PMC article. Review.
Cited by
- Palmitoylation of superoxide dismutase 1 (SOD1) is increased for familial amyotrophic lateral sclerosis-linked SOD1 mutants.
Antinone SE, Ghadge GD, Lam TT, Wang L, Roos RP, Green WN. Antinone SE, et al. J Biol Chem. 2013 Jul 26;288(30):21606-17. doi: 10.1074/jbc.M113.487231. Epub 2013 Jun 12. J Biol Chem. 2013. PMID: 23760509 Free PMC article. - Redox Mechanisms in Neurodegeneration: From Disease Outcomes to Therapeutic Opportunities.
Sbodio JI, Snyder SH, Paul BD. Sbodio JI, et al. Antioxid Redox Signal. 2019 Apr 10;30(11):1450-1499. doi: 10.1089/ars.2017.7321. Epub 2018 May 4. Antioxid Redox Signal. 2019. PMID: 29634350 Free PMC article. Review. - Oxidative stress in the development, maintenance and resolution of paclitaxel-induced painful neuropathy.
Duggett NA, Griffiths LA, McKenna OE, de Santis V, Yongsanguanchai N, Mokori EB, Flatters SJ. Duggett NA, et al. Neuroscience. 2016 Oct 1;333:13-26. doi: 10.1016/j.neuroscience.2016.06.050. Epub 2016 Jul 5. Neuroscience. 2016. PMID: 27393249 Free PMC article. - Copper trafficking in biology: an NMR approach.
Banci L, Bertini I, Ciofi-Baffoni S. Banci L, et al. HFSP J. 2009 Jun;3(3):165-75. doi: 10.2976/1.3078306. Epub 2009 Mar 18. HFSP J. 2009. PMID: 19949444 Free PMC article. - Altered thiol chemistry in human amyotrophic lateral sclerosis-linked mutants of superoxide dismutase 1.
Solsona C, Kahn TB, Badilla CL, Álvarez-Zaldiernas C, Blasi J, Fernandez JM, Alegre-Cebollada J. Solsona C, et al. J Biol Chem. 2014 Sep 26;289(39):26722-26732. doi: 10.1074/jbc.M114.565333. Epub 2014 Aug 4. J Biol Chem. 2014. PMID: 25096579 Free PMC article.
References
- Kikuchi H., Almer G., Yamashita S., Guegan C., Nagai M., Xu Z., Sosunov A.A., McKhann G.M., II, Przedborski S. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc. Natl Acad. Sci. USA. 2006;103:6025–6030. - PMC - PubMed
- Jaarsma D., Rognoni F., van Duijn W., Verspaget H.W., Haasdijk E.D., Holstege J.C. CuZn superoxide dismutase (SOD1) accumulates in vacuolated mitochondria in transgenic mice expressing amyotrophic lateral sclerosis-linked SOD1 mutations. Acta Neuropathol. (Berl.) 2001;102:293–305. - PubMed
- Okado-Matsumoto A., Fridovich I. Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J. Biol. Chem. 2001;276:38388–38393. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F31 NS054554/NS/NINDS NIH HHS/United States
- R01 NS051419-01A1/NS/NINDS NIH HHS/United States
- R01 NS051419-02/NS/NINDS NIH HHS/United States
- R01 NS051419/NS/NINDS NIH HHS/United States
- R01 NS051419-04/NS/NINDS NIH HHS/United States
- R01-NS051419/NS/NINDS NIH HHS/United States
- R01 NS051419-03/NS/NINDS NIH HHS/United States
- P01-NS011766/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous