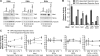TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription - PubMed (original) (raw)
TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription
Jer-Yuan Hsu et al. Genes Dev. 2008.
Abstract
The RNA polymerase II core promoter is a structurally and functionally diverse transcriptional module. RNAi depletion and overexpression experiments revealed a genetic circuit that controls the balance of transcription from two core promoter motifs, the TATA box and the downstream core promoter element (DPE). In this circuit, TBP activates TATA-dependent transcription and represses DPE-dependent transcription, whereas Mot1 and NC2 block TBP function and thus repress TATA-dependent transcription and activate DPE-dependent transcription. This regulatory circuit is likely to be one means by which biological networks can transmit transcriptional signals, such as those from DPE-specific and TATA-specific enhancers, via distinct pathways.
Figures
Figure 1.
Depletion of TBP reduces TATA-dependent but not DPE-dependent transcription. Drosophila S2 cells were depleted of the indicated factors by RNAi, and then transfected with TATA-dependent or DPE-dependent luciferase reporter genes. The activities of the RNAi-depleted extracts are reported as luminescent units per microgram of protein of RNAi-depleted extracts relative to the luminescent units per microgram of protein of mock RNAi-treated control extracts. The experimental scheme and reporter constructs are depicted at the bottom of the figure. “PC4-like” is Ssb-C31a, which is the Drosophila protein that is most closely related to mammalian PC4. The error bars represent the standard deviation.
Figure 2.
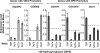
Mot1 and NC2 act in opposition to TBP to promote DPE transcription relative to TATA transcription. (A) Mot1, NC2, and TBP are efficiently depleted by RNAi in S2 cells. (B) The ability of Mot1 and NC2 to affect DPE-dependent versus TATA-dependent transcription requires TBP. RNAi depletion analysis of the indicated factors was carried out as in Figure 1. (C) Overexpression of TBP has the opposite effect as overexpression of Mot1 or NC2 upon DPE-dependent versus TATA-dependent transcription. The indicated expression vectors were cotransfected with DPE-dependent or TATA-dependent luciferase reporter genes. In each series of transfections, the total amount of expression vector was maintained at a constant level by the inclusion, where necessary, of a compensatory amount of empty vector (pAc5.1). The reporter gene activities with the expression vectors are given relative to those obtained with the empty vector alone. The error bars represent the standard deviation.
Figure 3.
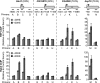
Mot1 and NC2 have opposite effects as TBP upon transcription of DPE- versus TATA-containing endogenous genes in Drosophila cells. These studies examined secondary/late ecdysone-responsive genes that are activated upon ecdysone induction. Drosophila Kc167 cells were depleted of the indicated factors by RNAi, and then induced with 20-hydroxyecdysone (20HE; 1 μM) for 24 h. The transcription levels of two TATA-containing genes (that lack DPE motifs) and three DPE-containing genes (that lack TATA elements) were determined by real-time RT–PCR. The error bars represent the SEM.
Figure 4.
TBP ChIP increases in TATA- but not DPE-containing promoters upon ecdysone induction. ChIP analyses of TBP and RNA polymerase II (Rpb3 subunit) were carried out with Drosophila Kc167 cells in the absence or presence of 20HE. Three ecdysone-responsive genes as well as hsp70, as a reference/control, were analyzed. The amounts of immunoprecipitated DNA with each set of the indicated primers were quantitated by real-time PCR. The enrichments observed with anti-TBP or anti-Rpb3 relative to a control nonimmune serum are shown. The error bars represent the SEM from three independent sets of experiments. The diagrams of the genes are not drawn to scale.
Figure 5.
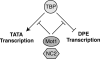
Mot1, NC2, and TBP create a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. In this model, TBP activates TATA transcription and represses DPE transcription, and Mot1 and NC2 act to inhibit the function of TBP. Thus, a decrease in TBP and/or an increase in Mot1/NC2 favors DPE transcription, whereas a decrease in Mot1/NC2 and/or an increase in TBP favors TBP transcription.
Similar articles
- Caudal, a key developmental regulator, is a DPE-specific transcriptional factor.
Juven-Gershon T, Hsu JY, Kadonaga JT. Juven-Gershon T, et al. Genes Dev. 2008 Oct 15;22(20):2823-30. doi: 10.1101/gad.1698108. Genes Dev. 2008. PMID: 18923080 Free PMC article. - Core promoter-specific gene regulation: TATA box selectivity and Initiator-dependent bi-directionality of serum response factor-activated transcription.
Xu M, Gonzalez-Hurtado E, Martinez E. Xu M, et al. Biochim Biophys Acta. 2016 Apr;1859(4):553-63. doi: 10.1016/j.bbagrm.2016.01.005. Epub 2016 Jan 26. Biochim Biophys Acta. 2016. PMID: 26824723 Free PMC article. - The DPE, a core promoter element for transcription by RNA polymerase II.
Kadonaga JT. Kadonaga JT. Exp Mol Med. 2002 Sep 30;34(4):259-64. doi: 10.1038/emm.2002.36. Exp Mol Med. 2002. PMID: 12515390 Review. - Mot1 associates with transcriptionally active promoters and inhibits association of NC2 in Saccharomyces cerevisiae.
Geisberg JV, Moqtaderi Z, Kuras L, Struhl K. Geisberg JV, et al. Mol Cell Biol. 2002 Dec;22(23):8122-34. doi: 10.1128/MCB.22.23.8122-8134.2002. Mol Cell Biol. 2002. PMID: 12417716 Free PMC article. - Role of the TATA-box binding protein (TBP) and associated family members in transcription regulation.
Mishal R, Luna-Arias JP. Mishal R, et al. Gene. 2022 Jul 30;833:146581. doi: 10.1016/j.gene.2022.146581. Epub 2022 May 18. Gene. 2022. PMID: 35597524 Review.
Cited by
- Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome.
van Werven FJ, van Teeffelen HA, Holstege FC, Timmers HT. van Werven FJ, et al. Nat Struct Mol Biol. 2009 Oct;16(10):1043-8. doi: 10.1038/nsmb.1674. Epub 2009 Sep 20. Nat Struct Mol Biol. 2009. PMID: 19767748 - TFIIB recognition elements control the TFIIA-NC2 axis in transcriptional regulation.
Deng W, Malecová B, Oelgeschläger T, Roberts SG. Deng W, et al. Mol Cell Biol. 2009 Mar;29(6):1389-400. doi: 10.1128/MCB.01346-08. Epub 2008 Dec 29. Mol Cell Biol. 2009. PMID: 19114554 Free PMC article. - The punctilious RNA polymerase II core promoter.
Vo Ngoc L, Wang YL, Kassavetis GA, Kadonaga JT. Vo Ngoc L, et al. Genes Dev. 2017 Jul 1;31(13):1289-1301. doi: 10.1101/gad.303149.117. Genes Dev. 2017. PMID: 28808065 Free PMC article. Review. - Caudal, a key developmental regulator, is a DPE-specific transcriptional factor.
Juven-Gershon T, Hsu JY, Kadonaga JT. Juven-Gershon T, et al. Genes Dev. 2008 Oct 15;22(20):2823-30. doi: 10.1101/gad.1698108. Genes Dev. 2008. PMID: 18923080 Free PMC article. - TRF2: TRansForming the view of general transcription factors.
Zehavi Y, Kedmi A, Ideses D, Juven-Gershon T. Zehavi Y, et al. Transcription. 2015;6(1):1-6. doi: 10.1080/21541264.2015.1004980. Epub 2015 Jan 14. Transcription. 2015. PMID: 25588059 Free PMC article. Review.
References
- Auble D.T., Hansen K.E., Mueller C.G.F., Lane W.S., Thorner J., Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes & Dev. 1994;8:1920–1934. - PubMed
- Buratowski S., Hahn S., Guarente L., Sharp P.A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. - PubMed
- Burke T.W., Kadonaga J.T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes & Dev. 1996;10:711–724. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases