Small CRISPR RNAs guide antiviral defense in prokaryotes - PubMed (original) (raw)
Small CRISPR RNAs guide antiviral defense in prokaryotes
Stan J J Brouns et al. Science. 2008.
Abstract
Prokaryotes acquire virus resistance by integrating short fragments of viral nucleic acid into clusters of regularly interspaced short palindromic repeats (CRISPRs). Here we show how virus-derived sequences contained in CRISPRs are used by CRISPR-associated (Cas) proteins from the host to mediate an antiviral response that counteracts infection. After transcription of the CRISPR, a complex of Cas proteins termed Cascade cleaves a CRISPR RNA precursor in each repeat and retains the cleavage products containing the virus-derived sequence. Assisted by the helicase Cas3, these mature CRISPR RNAs then serve as small guide RNAs that enable Cascade to interfere with virus proliferation. Our results demonstrate that the formation of mature guide RNAs by the CRISPR RNA endonuclease subunit of Cascade is a mechanistic requirement for antiviral defense.
Figures
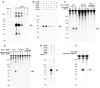
Fig. 1
The composition of the Cascade complex. (A) Schematic diagram of the CRISPR/cas gene cluster of E. coli K12 W3110. Repeats and spacers are indicated by diamonds and rectangles, respectively. A palindrome in the repeat is marked by convergently pointing arrows. Protein family nomenclature is as described in (11, 12). (B) Coomassie blue—stained SDS-polyacrylamide gel of the affinity purified protein complex using either the N-terminal StrepII-tag (S) or C-terminal His-tag (H) of each of the subunits CasB, CasC, CasD, or CasE as bait. Asterisks indicate the 5.5 kD larger double-tagged subunits. Marker sizes in kilodaltons on the left; location of untagged subunits on the right.
Fig. 2
Cascade cleaves CRISPR RNA precursors into small RNAs of ∼57 nucleotides (marked by arrows). (A) Northern analysis of total RNA of WT E. coli K12 (WT), a non-cas gene knockout (Δ_u, uidA_, β-glucuronidase), and Cascade gene knockouts using the single-stranded spacer sequence BG2349 (table S2) as a probe. (B) Northern blot as in (A) of total RNA from E. coli BL21 (DE3) expressing the E. coli K12 pre-crRNA and either the complete or incomplete Cascade complex. (C) Activity assays with purified Cascade using in vitro transcribed α-32P–uridine triphosphate–labeled pre-crRNA from E. coli K12 (repeat sequence: GAGUUCCCCGCCAGCGGGGAUAAACCG), E. coli UTI89 (repeat sequence: GUUCACUGCCGUACAGGCAGCUUAGAAA), and non-crRNA as substrates. (D) Activity assays as shown in (C) for 15 min with purified MalE-LacZα and MalE-CasE fusion proteins. (E) Northern blot as shown in (B) with Cascade or Cascade-CasEH20A. (F) Activity assays as shown in (C) for 30 min with purified Cascade or Cascade-CasEH20A.
Fig. 3
Cleaved crRNAs remain bound by Cascade. (A) Denaturing polyacryl-amide gel showing the crRNA (marked by the arrow) isolated from purified Cascade in the absence and presence of co-expressed pre-crRNA. (B) Secondary structure of pre-crRNA repeats and example sequences of cloned crRNAs indicating the PCS and crRNA handles.
Fig. 4
Engineered CRISPRs confer resistance to λ in the presence of Cascade and Cas3. (A) Effect of the presence of different sets of cas genes on the sensitivity of E. coli to phage λvir. Cells were equipped with one of two engineered CRISPRs containing four anti-λ spacers each (fig. S3). The C1–4 CRISPR produces crRNA complementary to the coding strand and mRNA of λvir, and the T1–4 CRISPR targets only the template strand. The sensitivity of each strain to phage λvir is represented as a histogram of the efficiency of plaquing, which is the plaque count ratio of the anti-λ CRISPR to that of the nontargeting control CRISPR. (B) Effect of single anti-λ spacers (fig. S3) on the sensitivity of E. coli to λvir. Error bars indicate 1 SD.
Comment in
- Molecular biology. Secret weapon.
Young RF 3rd. Young RF 3rd. Science. 2008 Aug 15;321(5891):922-3. doi: 10.1126/science.1162910. Science. 2008. PMID: 18703730 Free PMC article. No abstract available.
Similar articles
- CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference.
Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, Nogales E, Doudna JA. Hochstrasser ML, et al. Proc Natl Acad Sci U S A. 2014 May 6;111(18):6618-23. doi: 10.1073/pnas.1405079111. Epub 2014 Apr 18. Proc Natl Acad Sci U S A. 2014. PMID: 24748111 Free PMC article. - CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli.
Díez-Villaseñor C, Guzmán NM, Almendros C, García-Martínez J, Mojica FJ. Díez-Villaseñor C, et al. RNA Biol. 2013 May;10(5):792-802. doi: 10.4161/rna.24023. Epub 2013 Feb 27. RNA Biol. 2013. PMID: 23445770 Free PMC article. - Helicase dissociation and annealing of RNA-DNA hybrids by Escherichia coli Cas3 protein.
Howard JA, Delmas S, Ivančić-Baće I, Bolt EL. Howard JA, et al. Biochem J. 2011 Oct 1;439(1):85-95. doi: 10.1042/BJ20110901. Biochem J. 2011. PMID: 21699496 - The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity.
Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. Westra ER, et al. Annu Rev Genet. 2012;46:311-39. doi: 10.1146/annurev-genet-110711-155447. Annu Rev Genet. 2012. PMID: 23145983 Review. - Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes.
Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Al-Attar S, et al. Biol Chem. 2011 Apr;392(4):277-89. doi: 10.1515/BC.2011.042. Epub 2011 Feb 7. Biol Chem. 2011. PMID: 21294681 Review.
Cited by
- Opportunities for Riboswitch Inhibition by Targeting Co-Transcriptional RNA Folding Events.
Stephen C, Palmer D, Mishanina TV. Stephen C, et al. Int J Mol Sci. 2024 Sep 29;25(19):10495. doi: 10.3390/ijms251910495. Int J Mol Sci. 2024. PMID: 39408823 Free PMC article. Review. - CRISPR-Cas9 target-strand nicking provides phage resistance by inhibiting replication.
Nguyen GT, Schelling MA, Sashital DG. Nguyen GT, et al. bioRxiv [Preprint]. 2024 Sep 5:2024.09.05.611540. doi: 10.1101/2024.09.05.611540. bioRxiv. 2024. PMID: 39282300 Free PMC article. Preprint. - Cell-Penetrating Peptides and CRISPR-Cas9: A Combined Strategy for Human Genetic Disease Therapy.
de Morais CCPL, Correia EM, Bonamino MH, Vasconcelos ZFM. de Morais CCPL, et al. Hum Gene Ther. 2024 Oct;35(19-20):781-797. doi: 10.1089/hum.2024.020. Hum Gene Ther. 2024. PMID: 39276086 Review. - Synergistic effect of split DNA activators of Cas12a with exon-unwinding and induced targeting effect.
Huang S, Lou Y, Zheng L. Huang S, et al. Nucleic Acids Res. 2024 Oct 14;52(18):11148-11157. doi: 10.1093/nar/gkae766. Nucleic Acids Res. 2024. PMID: 39258555 Free PMC article.
References
- Barrangou R, et al. Science. 2007;315:1709. - PubMed
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. J Mol Evol. 2005;60:174. - PubMed
- Pourcel C, Salvignol G, Vergnaud G. Microbiology. 2005;151:653. - PubMed
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Microbiology. 2005;151:2551. - PubMed
- Godde JS, Bickerton A. J Mol Evol. 2006;62:718. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials