Quantitative analysis of redox-sensitive proteome with DIGE and ICAT - PubMed (original) (raw)
. 2008 Sep;7(9):3789-802.
doi: 10.1021/pr800233r. Epub 2008 Aug 16.
Affiliations
- PMID: 18707151
- PMCID: PMC2577071
- DOI: 10.1021/pr800233r
Quantitative analysis of redox-sensitive proteome with DIGE and ICAT
Cexiong Fu et al. J Proteome Res. 2008 Sep.
Abstract
Oxidative modifications of protein thiols are important mechanisms for regulating protein functions. The present study aimed to compare the relative effectiveness of two thiol-specific quantitative proteomic techniques, difference gel electrophoresis (DIGE) and isotope coded affinity tag (ICAT), for the discovery of redox-sensitive proteins in heart tissues. We found that these two methods were largely complementary; each could be used to reveal a set of unique redox-sensitive proteins. Some of these proteins are low-abundant signaling proteins and membrane proteins. From DIGE analysis, we found that both NF-kappaB-repressing protein and epoxide hydrolase were sensitive to H 2O 2 oxidation. In ICAT analysis, we found that specific cysteines within sacroplasmic endoplamic reticulum calcium ATPase 2 and voltage-dependent anion-selective channel protein 1 were sensitive to H 2O 2 oxidation. From these analyses, we conclude that both methods should be employed for proteome-wide studies, to maximize the possibility of identifying proteins containing redox-sensitive cysteinyl thiols in complex biological systems.
Figures
Figure 1
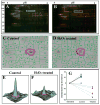
Identification of H2O2-sensitive heart proteins with saturation-labeling DIGE. Mouse heart proteins were incubated with H2O or 500 _μ_M H2O2 and labeled with Cy3m (green), each sample was mixed with an internal standard labeled with Cy5m (red). A mixture of a control sample and the internal standard was resolved in a 2D gel and the superimposed fluorescent image from the control and the standard proteome is shown in (A). The same procedure was applied to a H2O2-treated sample and the gel image is shown in (B). More green spots were observed in the control sample (A) than the H2O2 treated sample (B), when both are compared to the Cy5m labeled internal standard. This observation indicates that fewer free thiols were available for Cy3m labeling in the H2O2 treated sample (B). The white boxes in panels (A) and (B) highlight a group of succinate dehydrogenase isoforms that were sensitive to H2O2 oxidation (less green spots in B and confirmed with DeCyder quantification analysis). It is also noticeable that the acidic end of this group of protein isoforms appeared more red in A and greener in B, suggesting the appearance of more acidic isoforms under H2O2 challenge. Protein spots were detected and matched among the gels with the assistance of preassigned landmark spots. Consistent protein spot detection and matching is demonstrated between the control (C) and the H2O2-treated gels (D). The highlighted spots in panels (C) and (D) represent a protein (later identified as malate dehydrogenase) with significant decrease of fluorescent intensity following H2O2 treatment. This change is more strikingly illustrated in the 3D comparison of the spot volumes between a control (E) and a H2O2-treated sample (F). Quantitative analysis was carried out after spot volume normalization with the internal standard. Statistical evaluation (p < 0.005) (G) confirmed the significant decrease of ~70% of cysteine thiol in malate dehydrogenase following H2O2 treatment.
Figure 2
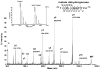
Example of ICAT identification of H2O2-sensitive heart proteins. A MS/MS spectrum for a peptide (204TIIPLISQCTPK215) from MDH isolated from mouse heart is presented with a continuous series of y-ions for confident identification. The observation of a mass difference of 339 Da between y3 and y4 ions matched the mass of a heavy ICAT reagent-modified cysteine. The decrease of cysteine thiol was observed in the MS spectrum (see insert). The hallmark of a pair of the heavy and light ICAT-labeled peptides is a mass difference of 9.03 Da between the two adjacent monoisotopic peaks in the MS spectrum. The relative ICAT ratio for this peptide was 0.73, indicating ~27% loss of the redox-sensitive cysteine thiols upon H2O2 treatment.
Figure 3
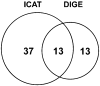
Venn diagram of H2O2-sensitive proteins discovered by DIGE and ICAT methods. We identified 50 proteins as potential targets of H2O2 oxidation by the ICAT method and 26 with the DIGE method. Of these, 13 proteins were identified with both methods. Overall, the ICAT technique enabled us to identify 24 more redox-sensitive proteins than the DIGE method.
Figure 4
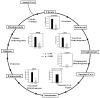
H2O2-sensitive TCA cycle proteins identified in this study. Both ICAT and DIGE methods revealed that many proteins in the TCA cycle were prone to oxidation by H2O2. The quantification of protein thiol level change with the ICAT method is the average of all its ICAT peptides. We detected a reduction of 20–30% of free cysteine thiols in aconitase 2, isocitrate dehydrogenase, succinate dehydrogenase, and malate dehydrogenase with the ICAT method, whereas a more dramatic reduction ranging from 70 to 90% for the same proteins was seen with the DIGE method. Succinyl-CoA ligase was observed solely by the ICAT method with a 35% free thiol level decrease, whereas oxoglutarate dehydrogenase oxidation was observed only with the DIGE method, with an over 95% decrease of free thiols. Student t test was carried out with four independent studies, *, p < 0.05; ***, p < 0.005.
Figure 5
DIGE analysis of MDH oxidized by H2O2. Cy3m (green)-labeled control sample and Cy5m (red)-labeled H2O2- treated porcine MDH. A series of MDH isoforms were observed with both untreated and H2O2-treated MDH. It was clear that the H2O2-treated MDH contained additional isoforms distributed toward the acidic end of the 2DE gel (red spots 1–3), suggesting the acidification of MDH, likely in the forms of cysteine oxidation to sulfinic, sulfenic, or sulfonic acids. The untreated MDH maintained isoforms clustered around the more basic p_I_ range, as illustrated by the green gel spots 6–7. The relative MDHreduction/MDHoxidation ratios for all MDH isoforms are marked below the corresponding gel spots, with isoform #1 being the most oxidized and isoform #7 being the most reduced. Ratio*: Cy3m/Cy5m ratio represents relative free cysteine thiol levels in control/H2O2-treated MDH isoforms.
Similar articles
- Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures.
Sethuraman M, McComb ME, Huang H, Huang S, Heibeck T, Costello CE, Cohen RA. Sethuraman M, et al. J Proteome Res. 2004 Nov-Dec;3(6):1228-33. doi: 10.1021/pr049887e. J Proteome Res. 2004. PMID: 15595732 - Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols.
Sethuraman M, McComb ME, Heibeck T, Costello CE, Cohen RA. Sethuraman M, et al. Mol Cell Proteomics. 2004 Mar;3(3):273-8. doi: 10.1074/mcp.T300011-MCP200. Epub 2004 Jan 15. Mol Cell Proteomics. 2004. PMID: 14726493 - The oxidized thiol proteome in fission yeast--optimization of an ICAT-based method to identify H2O2-oxidized proteins.
García-Santamarina S, Boronat S, Espadas G, Ayté J, Molina H, Hidalgo E. García-Santamarina S, et al. J Proteomics. 2011 Oct 19;74(11):2476-86. doi: 10.1016/j.jprot.2011.05.030. Epub 2011 Jun 6. J Proteomics. 2011. PMID: 21672643 - Concepts and approaches towards understanding the cellular redox proteome.
Ströher E, Dietz KJ. Ströher E, et al. Plant Biol (Stuttg). 2006 Jul;8(4):407-18. doi: 10.1055/s-2006-923961. Plant Biol (Stuttg). 2006. PMID: 16906481 Review. - Proteomic Characterization of Reversible Thiol Oxidations in Proteomes and Proteins.
Boronat S, Domènech A, Hidalgo E. Boronat S, et al. Antioxid Redox Signal. 2017 Mar 1;26(7):329-344. doi: 10.1089/ars.2016.6720. Epub 2016 May 20. Antioxid Redox Signal. 2017. PMID: 27089838 Review.
Cited by
- Identification of thioredoxin target protein networks in cardiac tissues of a transgenic mouse.
Fu C, Liu T, Parrott AM, Li H. Fu C, et al. Methods Mol Biol. 2013;1005:181-97. doi: 10.1007/978-1-62703-386-2_15. Methods Mol Biol. 2013. PMID: 23606258 Free PMC article. - Ethanol Attenuates Histiotrophic Nutrition Pathways and Alters the Intracellular Redox Environment and Thiol Proteome during Rat Organogenesis.
Jilek JL, Sant KE, Cho KH, Reed MS, Pohl J, Hansen JM, Harris C. Jilek JL, et al. Toxicol Sci. 2015 Oct;147(2):475-89. doi: 10.1093/toxsci/kfv145. Epub 2015 Jul 15. Toxicol Sci. 2015. PMID: 26185205 Free PMC article. - Upregulation of Nox4 by hypertrophic stimuli promotes apoptosis and mitochondrial dysfunction in cardiac myocytes.
Ago T, Kuroda J, Pain J, Fu C, Li H, Sadoshima J. Ago T, et al. Circ Res. 2010 Apr 16;106(7):1253-64. doi: 10.1161/CIRCRESAHA.109.213116. Epub 2010 Feb 25. Circ Res. 2010. PMID: 20185797 Free PMC article. - What can we learn about cardioprotection from the cardiac mitochondrial proteome?
Gucek M, Murphy E. Gucek M, et al. Cardiovasc Res. 2010 Nov 1;88(2):211-8. doi: 10.1093/cvr/cvq277. Epub 2010 Aug 30. Cardiovasc Res. 2010. PMID: 20805096 Free PMC article. Review. - S-nitrosylation: a radical way to protect the heart.
Murphy E, Kohr M, Sun J, Nguyen T, Steenbergen C. Murphy E, et al. J Mol Cell Cardiol. 2012 Mar;52(3):568-77. doi: 10.1016/j.yjmcc.2011.08.021. Epub 2011 Aug 27. J Mol Cell Cardiol. 2012. PMID: 21907718 Free PMC article. Review.
References
- Ghezzi P, Bonetto V. Redox proteomics: Identification of oxidatively, modified proteins. Proteomics. 2003;3(7):1145–53. - PubMed
- Di Simplicio P, Franconi F, Frosali S, Di Giuseppe D. Thiolation and nitrosation of cysteines in biological fluids and cells. Amino Acids. 2003;25(3–4):323–39. - PubMed
- Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279(5357):1718–21. - PubMed
- Abate C, Patel L, Rauscher FJ, Curran T. Redox regulation of fos and jun DNA-binding activity in vitro. Science. 1990;249(4973):1157–61. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials