OPCML is a broad tumor suppressor for multiple carcinomas and lymphomas with frequently epigenetic inactivation - PubMed (original) (raw)
. 2008 Aug 20;3(8):e2990.
doi: 10.1371/journal.pone.0002990.
Ying Ying, Andrew van Hasselt, Ka Man Ng, Jun Yu, Qian Zhang, Jie Jin, Dingxie Liu, Johng S Rhim, Sun Young Rha, Myriam Loyo, Anthony T C Chan, Gopesh Srivastava, George S W Tsao, Grant C Sellar, Joseph J Y Sung, David Sidransky, Qian Tao
Affiliations
- PMID: 18714356
- PMCID: PMC2500176
- DOI: 10.1371/journal.pone.0002990
OPCML is a broad tumor suppressor for multiple carcinomas and lymphomas with frequently epigenetic inactivation
Yan Cui et al. PLoS One. 2008.
Erratum in
- PLoS One. 2008;3(9). doi: 10.1371/annotation/f394b95b-c731-41a3-b0dc-be25fb6a227c
Abstract
Background: Identification of tumor suppressor genes (TSGs) silenced by CpG methylation uncovers the molecular mechanism of tumorigenesis and potential tumor biomarkers. Loss of heterozygosity at 11q25 is common in multiple tumors including nasopharyngeal carcinoma (NPC). OPCML, located at 11q25, is one of the downregulated genes we identified through digital expression subtraction.
Methodology/principal findings: Semi-quantitative RT-PCR showed frequent OPCML silencing in NPC and other common tumors, with no homozygous deletion detected by multiplex differential DNA-PCR. Instead, promoter methylation of OPCML was frequently detected in multiple carcinoma cell lines (nasopharyngeal, esophageal, lung, gastric, colon, liver, breast, cervix, prostate), lymphoma cell lines (non-Hodgkin and Hodgkin lymphoma, nasal NK/T-cell lymphoma) and primary tumors, but not in any non-tumor cell line and seldom weakly methylated in normal epithelial tissues. Pharmacological and genetic demethylation restored OPCML expression, indicating a direct epigenetic silencing. We further found that OPCML is stress-responsive, but this response is epigenetically impaired when its promoter becomes methylated. Ecotopic expression of OPCML led to significant inhibition of both anchorage-dependent and -independent growth of carcinoma cells with endogenous silencing.
Conclusions/significance: Thus, through functional epigenetics, we identified OPCML as a broad tumor suppressor, which is frequently inactivated by methylation in multiple malignancies.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
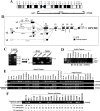
Figure 1. Identification of novel splicing variants of OPCML and its expression in normal human tissues.
(A) Genomic organization of the 11q25 locus with the two known genes OPCML and HNT. Transcriptional orientations are shown by curved arrows. (B) Different promoter usage and alternative splicing of OPCML. Alternative mRNA transcripts are shown aligned from 5′ to 3′ on a virtual genome. The 5′-end of _OPCML_-v1 assembled by ECgene (Genome Annotation for Alternative Splicing,
http://genome.ewha.ac.kr/ECgene/
) was adapted to this alignment. (C) Left panel: determination of transcription start sites of OPCML transcripts by 5′-RACE. Right panel: expression of _OPCML_-v1 and v3 in brain and testis by semi-quantitative RT-PCR using 5′-RACE product as the template. Primer pair v1F/R4 amplifies one band that is specific to v1. Primer pair v1F0/R4 amplifies two bands corresponding to v1 and v3, respectively. (D) Expression of _OPCML_-v1 and v3 in adult tissues by semi-quantitative RT-PCR. Primer pair v1F0/R4 amplifies two bands corresponding to v1 and v3, respectively. The specific and non-specific bands have been confirmed by direct sequencing. (E) Expression of _OPCML_-v1 and v2 in human normal adult and fetal tissues. Primer pair v1F/R4, v2F/R4 and F3/R2 are specific to the v1-, v2-transcripts, and common exons (exon 2 and 3) of OPCML , respectively. Sk.M., skeletal muscle. (F) Possible transcription of OPCML from other unidentified alternative promoters. Expression of OPCML in normal and tumor cell lines was analyzed by semi-quantitative RT-PCR using primers (F3/R2) specific to common exons (exon 2 and 3) of OPCML. Expression of _OPCML_-v1 or v2 is indicated as “+”, while downregulation or silencing is indicated as “−”. _OPCML_-v1 promoter methylation status in each cell line is also shown. M, methylated; U, unmethylated. Transcription of OPCML from unknown alternative promoters was found in some tumor cell lines (underlined) where the _OPCML_-v1 promoter is methylated and silenced and v2 expression is also silenced.
Figure 2. Epigenetic inactivation of OPCML in multiple tumor cell lines.
(A) Schematic structure of the v1 (NM_002545) CGI, with its exon 1, predicted promoter region, MSP region and BGS region indicated. Each short vertical line represents one CpG site. The v2 promoter is also shown. (B) Validation of the specificity of the MSP system. No signal was detected using the unbisulfited DNA from several tumor cell lines. (C) Representative analyses of OPCML v1 and v2 (NM_001012393) expression by semi-quantitative RT-PCR and methylation status of v1-CGI by MSP in tumor cell lines and normal controls. M, methylated; U, unmethylated. Immortalized normal epithelial cell lines (NE1, NE3, Het-1A, NP69, CCD 841, MLCSV40) and normal epithelial cell lines (HMEC, HMEpC and PrEC-6) with underlined names were used as normal controls. RC170N/h and RC165N/h are telomerase-immortalized benign prostate epithelial cell lines, RC92a/h and RC58T/h/SA#4 are telomerase-immortalized prostate tumor derived cell lines. (D) Expression and methylation of OPCML in glioma cell lines.
Figure 3. High-resolution methylation analysis of the _OPCML_-v1 promoter by BGS.
A region spanning the promoter with 90 CpG sites was analyzed. Each CpG site is shown at the top row as an individual number. Dense methylation of the v1-CGI was found in ESCC (EC18, EC109) and NPC (C666-1, CNE2, HK1) cell lines, but not in normal esophageal (NE1, NE3) and nasopharyngeal epithelial (NP69) cell lines. Five to 8 colonies of cloned BGS-PCR products from each bisulfite-treated DNA sample were sequenced and each is shown as an individual row, representing a single allele of the CGI analyzed. One circle indicates one CpG site. Dark filled or open circles represent methylated or unmethylated CpG sites, respectively. Δ indicates possible variation of a CpG site to the CpA or CpT dinucleotides. The MSP region in this study and the BGS region studied in the previous report are indicated in frames. The rightmost column is the MSP result of each sample.
Figure 4. Restorations of _OPCML_-v1 expression by demethylation.
(A) Pharmacological demethylation by Aza (A) and TSA (T) induced the expression of _OPCML_-v1 but not v2. OPCML expression before and after drug treatment was determined by RT-PCR. (B) Genetic demethylation of the _OPCML_-v1 CGI also activated its expression. _OPCML_-v1 expression in HCT116 cells and HCT116 with double knockout of DNMT1 and DNMT 3B (DKO) are shown. (C) Detailed BGS analysis confirmed the demethylation of the _OPCML_-v1 CGI in DKO cells.
Figure 5. Analysis of homozygous deletion of OPCML in multiple carcinoma cell lines and normal controls.
The abundance of OPCML relative to GAPDH was determined by multiplex differential genomic DNA PCR. The expression of OPCML in each sample is also shown. +, normal expression; −, downregulated/silenced.
Figure 6. OPCML-v1 was also methylated in different primary tumors.
(A) Frequent methylation of the _OPCML_-v1 CGI in multiple primary tumors as analyzed by MSP. M, methylated; U, unmethylated. Representative results are shown. T, tumors; N, paired non-tumor tissues. Good quality of bisulfited DNA samples of normal NPx (NPx1-4) has been confirmed by Q-MSP for beta-actin . (B) In contrast, no methylation was detected in prostate tumors. EsCa, esophageal carcinoma; HCC, hepatocellular carcinoma; GsCa, gastric carcinoma; BrCa, breast carcinoma; CxCa, cervical carcinoma.
Figure 7. The _OPCML_-v1 promoter is stress- and p53-responsive.
(A) Locations of transcription factors (HSF, p53, Sp1, E2F, STAT) binding sites in the promoter are indicated. (B) Up-regulation of _OPCML_-v1 in response to stress treatments is disrupted in tumor cell lines with a methylated promoter. Normal (NE3, HEK293, NE1) and tumor cell lines (Rael, CNE1, C666-1, HK1) were exposed to 42°C heat shock (HS), UV irradiation, or H2O2 treatments. _OPCML_-v1 promoter methylation status in each cell line is shown at the bottom. M, methylated; U, unmethylated. (C) H1299 cells were transfected with different amounts of pcDNA3.1+/TP53 (gift from Dr. Bert Vogelstein) . Expression of _OPCML_-v1 and v2 was analyzed by semi-quantitative RT-PCR. p53 induced a dosage-dependent upregulation of _OPCML_-v1.
Figure 8. Ectopic expression of OPCML-v1 inhibits tumor cell growth.
The effect of ectopic OPCML-v1 expression on carcinoma cell clonogenicity was investigated by monolayer colony formation assay (A) and soft agar assay (B). Cells were transfected with pcDNA3.1+/_OPCML_-v1 or control vector, and selected with G418. (C) Quantitative analyses of colony formation. The numbers of G418-resistant colonies in each vector-transfected control were set to 100%, while OPCML-v1 expressed cells were presented as mean±SD. Three independent experiments were performed in triplicate. The asterisk indicated statistical significant difference (p<0.01).
Similar articles
- Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation.
Ying J, Li H, Seng TJ, Langford C, Srivastava G, Tsao SW, Putti T, Murray P, Chan AT, Tao Q. Ying J, et al. Oncogene. 2006 Feb 16;25(7):1070-80. doi: 10.1038/sj.onc.1209154. Oncogene. 2006. PMID: 16247458 - OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer.
Sellar GC, Watt KP, Rabiasz GJ, Stronach EA, Li L, Miller EP, Massie CE, Miller J, Contreras-Moreira B, Scott D, Brown I, Williams AR, Bates PA, Smyth JF, Gabra H. Sellar GC, et al. Nat Genet. 2003 Jul;34(3):337-43. doi: 10.1038/ng1183. Nat Genet. 2003. PMID: 12819783 - Epigenetic identification of ADAMTS18 as a novel 16q23.1 tumor suppressor frequently silenced in esophageal, nasopharyngeal and multiple other carcinomas.
Jin H, Wang X, Ying J, Wong AH, Li H, Lee KY, Srivastava G, Chan AT, Yeo W, Ma BB, Putti TC, Lung ML, Shen ZY, Xu LY, Langford C, Tao Q. Jin H, et al. Oncogene. 2007 Nov 22;26(53):7490-8. doi: 10.1038/sj.onc.1210559. Epub 2007 Jun 4. Oncogene. 2007. PMID: 17546048 Free PMC article. - Emerging roles for the GPI-anchored tumor suppressor OPCML in cancers.
Antony J, Zanini E, Birtley JR, Gabra H, Recchi C. Antony J, et al. Cancer Gene Ther. 2021 Feb;28(1-2):18-26. doi: 10.1038/s41417-020-0187-6. Epub 2020 Jun 29. Cancer Gene Ther. 2021. PMID: 32595215 Review. - Tumor suppressor genes on frequently deleted chromosome 3p in nasopharyngeal carcinoma.
Chen J, Fu L, Zhang LY, Kwong DL, Yan L, Guan XY. Chen J, et al. Chin J Cancer. 2012 May;31(5):215-22. doi: 10.5732/cjc.011.10364. Epub 2012 Feb 24. Chin J Cancer. 2012. PMID: 22360856 Free PMC article. Review.
Cited by
- Epigenetic Biomarkers and the Wnt/β-Catenin Pathway in Opisthorchis viverrini-associated Cholangiocarcinoma: A Scoping Review on Therapeutic Opportunities.
Kafle A, Suttiprapa S, Muhammad M, Tenorio JCB, Mahato RK, Sahimin N, Loong SK. Kafle A, et al. PLoS Negl Trop Dis. 2024 Sep 5;18(9):e0012477. doi: 10.1371/journal.pntd.0012477. eCollection 2024 Sep. PLoS Negl Trop Dis. 2024. PMID: 39236081 Free PMC article. Review. - MicroRNAs Associated with IgLON Cell Adhesion Molecule Expression.
Salluzzo M, Vianello C, Flotta F, Rimondini R, Carboni L. Salluzzo M, et al. Curr Issues Mol Biol. 2024 Jul 19;46(7):7702-7718. doi: 10.3390/cimb46070456. Curr Issues Mol Biol. 2024. PMID: 39057097 Free PMC article. Review. - The Risk Genes for Neuropsychiatric Disorders negr1 and opcml Are Expressed throughout Zebrafish Brain Development.
Habicher J, Sanvido I, Bühler A, Sartori S, Piccoli G, Carl M. Habicher J, et al. Genes (Basel). 2024 Mar 14;15(3):363. doi: 10.3390/genes15030363. Genes (Basel). 2024. PMID: 38540422 Free PMC article. - The Role of IgLON Cell Adhesion Molecules in Neurodegenerative Diseases.
Salluzzo M, Vianello C, Abdullatef S, Rimondini R, Piccoli G, Carboni L. Salluzzo M, et al. Genes (Basel). 2023 Sep 28;14(10):1886. doi: 10.3390/genes14101886. Genes (Basel). 2023. PMID: 37895235 Free PMC article. Review. - Establishing a Prediction Model for the Efficacy of Platinum-Based Chemotherapy in NSCLC Based on a Two Cohorts GWAS Study.
Xiao Q, Mao C, Gao Y, Huang H, Yu B, Yu L, Li X, Mao X, Zhang W, Yin J, Liu Z. Xiao Q, et al. J Clin Med. 2023 Feb 7;12(4):1318. doi: 10.3390/jcm12041318. J Clin Med. 2023. PMID: 36835855 Free PMC article.
References
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. - PubMed
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. - PubMed
- Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. - PubMed
- Tao Q, Chan AT. Nasopharyngeal carcinoma: molecular pathogenesis and therapeutic developments. Expert Rev Mol Med. 2007;9:1–24. - PubMed
- Raab-Traub N. Epstein-Barr virus in the pathogenesis of NPC. Semin Cancer Biol. 2002;12:431–441. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical