The origins of 168, W23, and other Bacillus subtilis legacy strains - PubMed (original) (raw)
The origins of 168, W23, and other Bacillus subtilis legacy strains
Daniel R Zeigler et al. J Bacteriol. 2008 Nov.
Abstract
Bacillus subtilis is both a model organism for basic research and an industrial workhorse, yet there are major gaps in our understanding of the genomic heritage and provenance of many widely used strains. We analyzed 17 legacy strains dating to the early years of B. subtilis genetics. For three--NCIB 3610T, PY79, and SMY--we performed comparative genome sequencing. For the remainder, we used conventional sequencing to sample genomic regions expected to show sequence heterogeneity. Sequence comparisons showed that 168, its siblings (122, 160, and 166), and the type strains NCIB 3610 and ATCC 6051 are highly similar and are likely descendants of the original Marburg strain, although the 168 lineage shows genetic evidence of early domestication. Strains 23, W23, and W23SR are identical in sequence to each other but only 94.6% identical to the Marburg group in the sequenced regions. Strain 23, the probable W23 parent, likely arose from a contaminant in the mutagenesis experiments that produced 168. The remaining strains are all genomic hybrids, showing one or more "W23 islands" in a 168 genomic backbone. Each traces its origin to transformations of 168 derivatives with DNA from 23 or W23. The common prototrophic lab strain PY79 possesses substantial W23 islands at its trp and sac loci, along with large deletions that have reduced its genome 4.3%. SMY, reputed to be the parent of 168, is actually a 168-W23 hybrid that likely shares a recent ancestor with PY79. These data provide greater insight into the genomic history of these B. subtilis legacy strains.
Figures
FIG. 1.
DNA sequence heterogeneity in the trp region of B. subtilis legacy strains. The arrows show reading frames in the B. subtilis 168 genome, while the axis indicates the corresponding position in the genome sequence. For each strain, boxes indicate the sizes and positions of contiguous DNA sequences determined in this study. Open boxes show sequences that are essentially identical to that of strain 168, while hatched boxes show sequences identical to that of W23. The upward-facing triangle (▴) shows the position of the trpC2 deletion, while the inverted triangle (▾) shows the position of the gudB duplication. The strain group labeled “168,3610T” also includes ATCC 6051T and the Burkholder and Giles mutants 122, 160, and 166.
FIG. 2.
Visualization of DNA sequence heterogeneity revealed by genome resequencing of B. subtilis strains SMY (A) and PY79 (B). For each figure, four panels are shown aligned to the position in the B. subtilis genome sequence. Bar lines in the first row indicate called SNPs, as follows: black bars, nonsense SNPs; blue bars, missense SNPs; light blue bars, silent (no amino acid change) SNPs; turquoise bars, SNPs within intergenic region. Blue bars in the second row show uncalled ROI. Gold lines in the third row show fully resequenced residues, and red lines in the fourth row show the net difference (arbitrary units) in hybridization intensity between the two tested strains, with the indicated “peaks” indicating large hybridization differences. For PY79, the positions of areas 1 to 6 are marked.
FIG. 3.
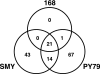
SNPs shared among B. subtilis 168, SMY, and PY79. Numbers indicate which SNPs are unique to a strain, which are shared between two strains, and which are shared among all three strains. The wild-type Marburg strain NCIB3610T was chosen as the root sequence for this comparison. Residues falling within “W23 islands” were excluded from the comparison.
FIG. 4.
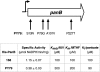
Correlation between sequence divergence and enzyme activity of PanB. The top panel shows the positions of the amino acid differences between the PanB enzymes from 168 and PY79; the bent arrow represents the wild-type sigma A promoter that controls transcription of panB. The bottom panel compares the activities of the His6-tagged PanB enzymes isolated from B. subtilis 168 and PY79 (with the latter containing the 29-kb B. subtilis W23-derived DNA island surrounding the trp locus). The values are means for three independent experiments.
FIG. 5.
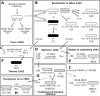
Model describing the history of early B. subtilis Trp+ legacy strains. The genomic heritage of individual strains is represented by shading, as follows: white cells, 168-like genomes; black cells, W23-like genomes; gray cells, 168-W23 hybrid genomes. The event leading to the isolation of a legacy strain is represented by a straight arrow (transformation, transduction, domestication, contamination, or renaming), zigzag arrow (mutagenesis through radiation), or dashed arrow (inferred but undocumented event). Rectangular boxes surround isolation events that are documented in the publications indicated.
Similar articles
- The genome sequence of Bacillus subtilis subsp. spizizenii W23: insights into speciation within the B. subtilis complex and into the history of B. subtilis genetics.
Zeigler DR. Zeigler DR. Microbiology (Reading). 2011 Jul;157(Pt 7):2033-2041. doi: 10.1099/mic.0.048520-0. Epub 2011 Apr 28. Microbiology (Reading). 2011. PMID: 21527469 - Sequencing and functional analysis of the genome of Bacillus subtilis strain 168.
Harwood CR, Wipat A. Harwood CR, et al. FEBS Lett. 1996 Jun 24;389(1):84-7. doi: 10.1016/0014-5793(96)00524-8. FEBS Lett. 1996. PMID: 8682212 Review. - Exploitation of Bacillus subtilis as a robust workhorse for production of heterologous proteins and beyond.
Cui W, Han L, Suo F, Liu Z, Zhou L, Zhou Z. Cui W, et al. World J Microbiol Biotechnol. 2018 Sep 10;34(10):145. doi: 10.1007/s11274-018-2531-7. World J Microbiol Biotechnol. 2018. PMID: 30203131 Review.
Cited by
- MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis.
Meeske AJ, Sham LT, Kimsey H, Koo BM, Gross CA, Bernhardt TG, Rudner DZ. Meeske AJ, et al. Proc Natl Acad Sci U S A. 2015 May 19;112(20):6437-42. doi: 10.1073/pnas.1504967112. Epub 2015 Apr 27. Proc Natl Acad Sci U S A. 2015. PMID: 25918422 Free PMC article. - Death Becomes Them: Bacterial Community Dynamics and Stilbene Antibiotic Production in Cadavers of Galleria mellonella Killed by Heterorhabditis and Photorhabdus spp.
Wollenberg AC, Jagdish T, Slough G, Hoinville ME, Wollenberg MS. Wollenberg AC, et al. Appl Environ Microbiol. 2016 Sep 16;82(19):5824-37. doi: 10.1128/AEM.01211-16. Print 2016 Oct 1. Appl Environ Microbiol. 2016. PMID: 27451445 Free PMC article. - Antibacterial Activity of Bacillus inaquosorum Strain T1 against pirABVp -Bearing Vibrio parahaemolyticus: Genetic and Physiological Characterization.
Avery SE, Ruzbarsky SP, Hise AM, Schreier HJ. Avery SE, et al. Appl Environ Microbiol. 2020 Oct 15;86(21):e01950-20. doi: 10.1128/AEM.01950-20. Print 2020 Oct 15. Appl Environ Microbiol. 2020. PMID: 32859595 Free PMC article. - Surfactin Shows Relatively Low Antimicrobial Activity against Bacillus subtilis and Other Bacterial Model Organisms in the Absence of Synergistic Metabolites.
Lilge L, Ersig N, Hubel P, Aschern M, Pillai E, Klausmann P, Pfannstiel J, Henkel M, Morabbi Heravi K, Hausmann R. Lilge L, et al. Microorganisms. 2022 Apr 5;10(4):779. doi: 10.3390/microorganisms10040779. Microorganisms. 2022. PMID: 35456828 Free PMC article. - Efficient plasmid transfer via natural competence in a microbial co-culture.
Cheng YY, Zhou Z, Papadopoulos JM, Zuke JD, Falbel TG, Anantharaman K, Burton BM, Venturelli OS. Cheng YY, et al. Mol Syst Biol. 2023 Mar 9;19(3):e11406. doi: 10.15252/msb.202211406. Epub 2023 Jan 30. Mol Syst Biol. 2023. PMID: 36714980 Free PMC article.
References
- Aertsen, A., I. Van Opstal, S. C. Vanmuysen, E. Y. Wuytack, and C. W. Michiels. 2005. Screening for Bacillus subtilis mutants deficient in pressure induced spore germination: identification of ykvU as a novel germination gene. FEMS Microbiol. Lett. 243385-391. - PubMed
- Albert, T. J., D. Dailidiene, G. Dailide, J. E. Norton, A. Kalia, T. A. Richmond, M. Molla, J. Singh, R. D. Green, and D. E. Berg. 2005. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat. Methods 2951-953. - PubMed
- Albertini, A. M., and A. Galizzi. 1999. The sequence of the trp operon of Bacillus subtilis 168 (trpC2) revisited. Microbiology 1453319-3320. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous