Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli - PubMed (original) (raw)
Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli
Junmei Zhang et al. Mol Cell Proteomics. 2009 Feb.
Abstract
Lysine acetylation and its regulatory enzymes are known to have pivotal roles in mammalian cellular physiology. However, the extent and function of this modification in prokaryotic cells remain largely unexplored, thereby presenting a hurdle to further functional study of this modification in prokaryotic systems. Here we report the first global screening of lysine acetylation, identifying 138 modification sites in 91 proteins from Escherichia coli. None of the proteins has been previously associated with this modification. Among the identified proteins are transcriptional regulators, as well as others with diverse functions. Interestingly, more than 70% of the acetylated proteins are metabolic enzymes and translation regulators, suggesting an intimate link of this modification to energy metabolism. The new dataset suggests that lysine acetylation could be abundant in prokaryotic cells. In addition, these results also imply that functions of lysine acetylation beyond regulation of gene expression are evolutionarily conserved from bacteria to mammals. Furthermore, we demonstrate that bacterial lysine acetylation is regulated in response to stress stimuli.
Figures
Fig. 1.
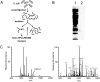
Synopsis of lysine acetylation proteomics. A, schematic representation of the sequential steps used for global profiling of lysine acetylation in E. coli. B, Western blotting analysis of protein lysate from E. coli with anti-acetyllysine antibody from ImmuneChem Inc. Lane 1, 30 μg of whole cell lysate from E. coli probed with anti-acetyllysine antibody; lane 2, competition with acetylated BSA (300 μg/ml). C, an example of MS and MS/MS analysis of a lysine-acetylated peptide for peptide identification and mapping of acetyllysine site in E. coli. Left panel, full MS spectrum at a retention time of 63.69 min. Right panel, MS/MS spectrum of m/z 1261.2, which identifies acetylated peptide YYQGTPSPVK*HPELTDMVIFR in isocitrate dehydrogenase, a metabolic protein in E. coli.
Fig. 2.
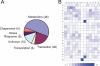
A, pie chart of functionally annotated protein groups that are lysine-acetylated. B, density map of lysine-acetylated peptides. The frequency of occurrence of amino acid residues surrounding sites of lysine acetylation was calculated, relative to the frequency of the residue within the entire E. coli genome, and schematically represented by a density map using a method described previously (11). Prevalence of specific amino acids at positions surrounding lysine acetylation sites is shown.
Fig. 3.
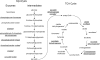
Lysine-acetylated proteins involved in glucose degradation and TCA cycle. Proteins identified as lysine-acetylated in the E. coli screen are underlined. Those identified in a screen of mammalian cells (11) are marked with an asterisk.
Fig. 4.
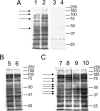
Western blotting analysis of protein lysate with an anti-acetyllysine antibody from ImmuneChem Inc. The 40-μg protein lysate was resolved in 4–20% SDS-PAGE. The Western blotting analysis was carried out as described previously (11). Each of the gel bands that have significant differences between the two E. coli strains or among the four oxygen conditions is labeled with an arrow on the left. The molecular weight markers are labeled on the right. A, lysine acetylation profiles of two E. coli strains (Lanes 1 and 3, MG1655; Lanes 2 and 4, JW1106). Lanes 1 and 2, probed with anti-acetyllysine antibody and non-acetylated BSA (300 μg/2 ml); Lanes 3 and 4, probed with anti-acetyllysine antibody and acetylated BSA (300 μg/2 ml). B, impact of nutritional conditions (E. coli MG1655). Lane 5, control and Lane 6, starvation. C, influences of oxygen conditions (E. coli MG1655). Lane 7, control (100% air); Lane 8, 25% air; Lane 9, 5% air; and Lane 10, 100% nitrogen.
Similar articles
- Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli.
Colak G, Xie Z, Zhu AY, Dai L, Lu Z, Zhang Y, Wan X, Chen Y, Cha YH, Lin H, Zhao Y, Tan M. Colak G, et al. Mol Cell Proteomics. 2013 Dec;12(12):3509-20. doi: 10.1074/mcp.M113.031567. Epub 2013 Oct 31. Mol Cell Proteomics. 2013. PMID: 24176774 Free PMC article. - The diversity of lysine-acetylated proteins in Escherichia coli.
Yu BJ, Kim JA, Moon JH, Ryu SE, Pan JG. Yu BJ, et al. J Microbiol Biotechnol. 2008 Sep;18(9):1529-36. J Microbiol Biotechnol. 2008. PMID: 18852508 - Ancient Regulatory Role of Lysine Acetylation in Central Metabolism.
Nakayasu ES, Burnet MC, Walukiewicz HE, Wilkins CS, Shukla AK, Brooks S, Plutz MJ, Lee BD, Schilling B, Wolfe AJ, Müller S, Kirby JR, Rao CV, Cort JR, Payne SH. Nakayasu ES, et al. mBio. 2017 Nov 28;8(6):e01894-17. doi: 10.1128/mBio.01894-17. mBio. 2017. PMID: 29184018 Free PMC article. - Studying lysine acetylation of citric acid cycle enzymes by genetic code expansion.
Fatema N, Fan C. Fatema N, et al. Mol Microbiol. 2023 May;119(5):551-559. doi: 10.1111/mmi.15052. Epub 2023 Mar 13. Mol Microbiol. 2023. PMID: 36890576 Free PMC article. Review. - Lysine acetylation and cancer: A proteomics perspective.
Gil J, Ramírez-Torres A, Encarnación-Guevara S. Gil J, et al. J Proteomics. 2017 Jan 6;150:297-309. doi: 10.1016/j.jprot.2016.10.003. Epub 2016 Oct 13. J Proteomics. 2017. PMID: 27746255 Review.
Cited by
- Mechanisms of transcription factor acetylation and consequences in hearts.
Thiagarajan D, Vedantham S, Ananthakrishnan R, Schmidt AM, Ramasamy R. Thiagarajan D, et al. Biochim Biophys Acta. 2016 Dec;1862(12):2221-2231. doi: 10.1016/j.bbadis.2016.08.011. Epub 2016 Aug 17. Biochim Biophys Acta. 2016. PMID: 27543804 Free PMC article. Review. - Lysine Acetylation and Succinylation in HeLa Cells and their Essential Roles in Response to UV-induced Stress.
Xu H, Chen X, Xu X, Shi R, Suo S, Cheng K, Zheng Z, Wang M, Wang L, Zhao Y, Tian B, Hua Y. Xu H, et al. Sci Rep. 2016 Jul 25;6:30212. doi: 10.1038/srep30212. Sci Rep. 2016. PMID: 27452117 Free PMC article. - Differential lysine acetylation profiles of Erwinia amylovora strains revealed by proteomics.
Wu X, Vellaichamy A, Wang D, Zamdborg L, Kelleher NL, Huber SC, Zhao Y. Wu X, et al. J Proteomics. 2013 Feb 21;79:60-71. doi: 10.1016/j.jprot.2012.12.001. Epub 2012 Dec 9. J Proteomics. 2013. PMID: 23234799 Free PMC article. - High-Resolution Metabolomics with Acyl-CoA Profiling Reveals Widespread Remodeling in Response to Diet.
Liu X, Sadhukhan S, Sun S, Wagner GR, Hirschey MD, Qi L, Lin H, Locasale JW. Liu X, et al. Mol Cell Proteomics. 2015 Jun;14(6):1489-500. doi: 10.1074/mcp.M114.044859. Epub 2015 Mar 20. Mol Cell Proteomics. 2015. PMID: 25795660 Free PMC article. - Succinylome analysis reveals the involvement of lysine succinylation in metabolism in pathogenic Mycobacterium tuberculosis.
Yang M, Wang Y, Chen Y, Cheng Z, Gu J, Deng J, Bi L, Chen C, Mo R, Wang X, Ge F. Yang M, et al. Mol Cell Proteomics. 2015 Apr;14(4):796-811. doi: 10.1074/mcp.M114.045922. Epub 2015 Jan 20. Mol Cell Proteomics. 2015. PMID: 25605462 Free PMC article.
References
- Haigis, M. C., and Guarente, L. P. ( 2006) Mammalian sirtuins-emerging roles in physiology, aging, and calorie restriction. Genes Dev. 20, 2913–2921 - PubMed
- McKinsey, T. A., and Olson, E. N. ( 2004) Cardiac histone acetylation – therapeutic opportunities abound. Trends Genet. 20, 206–213 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 RR020839/RR/NCRR NIH HHS/United States
- U45 RR020839/RR/NCRR NIH HHS/United States
- CA107943/CA/NCI NIH HHS/United States
- R33 CA107943/CA/NCI NIH HHS/United States
- R21 CA107943/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases