Identification of ALK as a major familial neuroblastoma predisposition gene - PubMed (original) (raw)
. 2008 Oct 16;455(7215):930-5.
doi: 10.1038/nature07261. Epub 2008 Aug 24.
Marci Laudenslager, Luca Longo, Kristina A Cole, Andrew Wood, Edward F Attiyeh, Michael J Laquaglia, Rachel Sennett, Jill E Lynch, Patrizia Perri, Geneviève Laureys, Frank Speleman, Cecilia Kim, Cuiping Hou, Hakon Hakonarson, Ali Torkamani, Nicholas J Schork, Garrett M Brodeur, Gian P Tonini, Eric Rappaport, Marcella Devoto, John M Maris
Affiliations
- PMID: 18724359
- PMCID: PMC2672043
- DOI: 10.1038/nature07261
Identification of ALK as a major familial neuroblastoma predisposition gene
Yaël P Mossé et al. Nature. 2008.
Abstract
Neuroblastoma is a childhood cancer that can be inherited, but the genetic aetiology is largely unknown. Here we show that germline mutations in the anaplastic lymphoma kinase (ALK) gene explain most hereditary neuroblastomas, and that activating mutations can also be somatically acquired. We first identified a significant linkage signal at chromosome bands 2p23-24 using a whole-genome scan in neuroblastoma pedigrees. Resequencing of regional candidate genes identified three separate germline missense mutations in the tyrosine kinase domain of ALK that segregated with the disease in eight separate families. Resequencing in 194 high-risk neuroblastoma samples showed somatically acquired mutations in the tyrosine kinase domain in 12.4% of samples. Nine of the ten mutations map to critical regions of the kinase domain and were predicted, with high probability, to be oncogenic drivers. Mutations resulted in constitutive phosphorylation, and targeted knockdown of ALK messenger RNA resulted in profound inhibition of growth in all cell lines harbouring mutant or amplified ALK, as well as in two out of six wild-type cell lines for ALK. Our results demonstrate that heritable mutations of ALK are the main cause of familial neuroblastoma, and that germline or acquired activation of this cell-surface kinase is a tractable therapeutic target for this lethal paediatric malignancy.
Figures
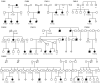
Figure 1. Eight neuroblastoma pedigrees with ALK mutations
All family members with DNA available for genotyping indicated with either wild type (wt) for ALK, or with mutation in the ALK tyrosine kinase domain (R1192P, R1275Q, G1128A). Individuals affected by neuroblastoma indicated by filled symbol.
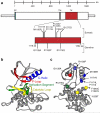
Figure 2. Germline and somatic ALK mutations
a. Schematic indicating protein structure of ALK, with mutations discovered in constitutional DNAs of familial cases (germline) and primary tumors from sporadic cases (somatic) indicated. All but one sequence alteration mapped to the tyrosine kinase domain (D1091N was just N-terminal and is not indicated here). Of the three germline mutations discovered, only the R1275Q was found in the tumor DNA samples. Conversely, the I1250T mutation discovered in the tumor set was also present in the matched germline DNA of that patient, while all of the other mutations studied here were somatically acquired. b. Homology model of wild-type ALK with each major subdomain indicated, . c. ALK mutations mapped onto homology model (orientation different to show all mutations) with colors indicating subdomain in which the mutation resides (e.g. the R1275Q mutation falls within the activation segment, indicated in green).
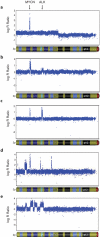
Figure 3. Representative ALK copy number alterations in three neuroblastoma primary tumors
Hybridization intensity reflecting copy number for all SNPs along chromosome 2p in three primary tumors from patients with sporadically occurring disease is shown, represented on a logarithmic scale. MYCN amplification is present in all tumors. a. Regional gain (trisomy) of chromosome 2p, including the ALK locus. b. Focal gain of the ALK locus. c. Focal amplification of the ALK locus. d and e. Complex rearrangement of the 2p locus, showing various focal amplicons, including MYCN and ALK.
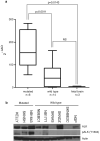
Figure 4. ALK is highly expressed and the kinase is phosphorylated in neuroblastoma cell lines harboring activating mutations
a. Relative ALK expression of neuroblastoma cell lines and fetal brain is shown and was determined using the 2-ΔΔCt method. Statistical significance was determined by unpaired T-test. b. Immunoblots showing differential ALK expression in neuroblastoma cell lines with phosphorylation of the tyrosine 1604 codon restricted to cell lines with mutations (the wild-type lines NBEC1 and NB1771 show faint phosphostaining).
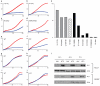
Figure 5. Inhibition of growth in ALK mutated or amplified neuroblastoma cell lines
Substrate adherent cellular growth was monitored and recorded every 30 minutes for at least 120 hours in ten neuroblastoma cell lines that were untreated, transfected with siRNAs against ALK, GAPDH (negative control), a non-targeting control (NTC, negative control) or the polo-like kinase gene (PLK1; positive control). The x-axis is time in hours after transfection, and differs based on the growth kinetics of each cell line. The y-axis is percent growth normalized to the siRNA against GAPDH. The normalized average percent growth is shown for siRNAs against GAPDH (red) and ALK (blue). In each experiment the siRNA NTC did not differ significantly from the siRNA against GAPDH, and in each case the siRNA against PLK1 showed profound growth inhibition, but only the siRNA against GAPDH and ALK are shown here for ease of visualization. Each experiment was plated in triplicate, and each experiment was repeated at least once with concordant results (data not shown). The replicates for each siRNA curve showed little variation with standard deviations <5% in all lines (except the siRNA against GAPDH in SKNDZ and NB1643 which were <10%). The cell lines, their mutation status and percent ALK mRNA knockdown at 48 hours are as follows: a. KELLY (F1174L) 78% mRNA knockdown (kd); b. SKNSH (F1174L), 21% mRNA kd; c. NB1643 (R1275Q), 48% mRNA kd; d. IMR5 (AMPLIFIED), 60% mRNA kd; e. NBEBC1 (WT), 74% mRNA kd; f. NB1771 (WT), 68% mRNA kd; g. SKNAS (WT), ALK not expressed at detectable levels; h. SKNDZ (WT), 41% mRNA kd; i. SKNBE2C (WT), 62% mRNA kd; j. NGP (WT), 86% mRNA kd. k. Summary of percentage growth inhibition with ALK siRNA knockdown by ALK mutational and allelic status; l. Immunoblot showing ALK protein knockdown at 24, 48 and 72 hours following siRNA transfection in the representative neuroblastoma cell lines KELLY (F1174L mutation and showing growth inhibition) and SKNDZ (wild-type ALK sequence and no growth inhibition).
Comment in
- Cancer: A ringleader identified.
Eng C. Eng C. Nature. 2008 Oct 16;455(7215):883-4. doi: 10.1038/455883a. Nature. 2008. PMID: 18923503 No abstract available.
Similar articles
- Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma.
Janoueix-Lerosey I, Lequin D, Brugières L, Ribeiro A, de Pontual L, Combaret V, Raynal V, Puisieux A, Schleiermacher G, Pierron G, Valteau-Couanet D, Frebourg T, Michon J, Lyonnet S, Amiel J, Delattre O. Janoueix-Lerosey I, et al. Nature. 2008 Oct 16;455(7215):967-70. doi: 10.1038/nature07398. Nature. 2008. PMID: 18923523 - Oncogenic mutations of ALK kinase in neuroblastoma.
Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, Wang L, Soda M, Kikuchi A, Igarashi T, Nakagawara A, Hayashi Y, Mano H, Ogawa S. Chen Y, et al. Nature. 2008 Oct 16;455(7215):971-4. doi: 10.1038/nature07399. Nature. 2008. PMID: 18923524 - Oncogenic mutations of ALK in neuroblastoma.
Ogawa S, Takita J, Sanada M, Hayashi Y. Ogawa S, et al. Cancer Sci. 2011 Feb;102(2):302-8. doi: 10.1111/j.1349-7006.2010.01825.x. Cancer Sci. 2011. PMID: 21205076 Free PMC article. Review. - Activating mutations in ALK provide a therapeutic target in neuroblastoma.
George RE, Sanda T, Hanna M, Fröhling S, Luther W 2nd, Zhang J, Ahn Y, Zhou W, London WB, McGrady P, Xue L, Zozulya S, Gregor VE, Webb TR, Gray NS, Gilliland DG, Diller L, Greulich H, Morris SW, Meyerson M, Look AT. George RE, et al. Nature. 2008 Oct 16;455(7215):975-8. doi: 10.1038/nature07397. Nature. 2008. PMID: 18923525 Free PMC article. - The emerging pathogenic and therapeutic importance of the anaplastic lymphoma kinase gene.
Kelleher FC, McDermott R. Kelleher FC, et al. Eur J Cancer. 2010 Sep;46(13):2357-68. doi: 10.1016/j.ejca.2010.04.006. Epub 2010 May 5. Eur J Cancer. 2010. PMID: 20451371 Review.
Cited by
- Identification of novel markers for neuroblastoma immunoclustering using machine learning.
Zhang L, Li H, Sun F, Wu Q, Jin L, Xu A, Chen J, Yang R. Zhang L, et al. Front Immunol. 2024 Nov 4;15:1446273. doi: 10.3389/fimmu.2024.1446273. eCollection 2024. Front Immunol. 2024. PMID: 39559348 Free PMC article. - Targeting KDM1A in Neuroblastoma with NCL-1 Induces a Less Aggressive Phenotype and Suppresses Angiogenesis.
Sprüssel A, Suzuki T, Miyata N, Astrahantseff K, Szymansky A, Toedling J, Thole-Kliesch TM, Ballagee A, Lodrini M, Künkele A, Truss M, Heukamp LC, Mathia S, Hertwig F, Rosenberger C, Eggert A, Deubzer HE, Schulte JH. Sprüssel A, et al. J Clin Med. 2024 Oct 12;13(20):6081. doi: 10.3390/jcm13206081. J Clin Med. 2024. PMID: 39458030 Free PMC article. - The Use of Anaplastic Lymphoma Kinase Inhibitors in Non-Small-Cell Lung Cancer Treatment-Literature Review.
Gorzelak-Magiera A, Domagała-Haduch M, Kabut J, Gisterek-Grocholska I. Gorzelak-Magiera A, et al. Biomedicines. 2024 Oct 11;12(10):2308. doi: 10.3390/biomedicines12102308. Biomedicines. 2024. PMID: 39457620 Free PMC article. Review. - A proteogenomic surfaceome study identifies DLK1 as an immunotherapeutic target in neuroblastoma.
Hamilton AK, Radaoui AB, Tsang M, Martinez D, Conkrite KL, Patel K, Sidoli S, Delaidelli A, Modi A, Rokita JL, Lane MV, Hartnett N, Lopez RD, Zhang B, Zhong C, Ennis B, Miller DP, Brown MA, Rathi KS, Raman P, Pogoriler J, Bhatti T, Pawel B, Glisovic-Aplenc T, Teicher B, Erickson SW, Earley EJ, Bosse KR, Sorensen PH, Krytska K, Mosse YP, Havenith KE, Zammarchi F, van Berkel PH, Smith MA, Garcia BA, Maris JM, Diskin SJ. Hamilton AK, et al. Cancer Cell. 2024 Nov 11;42(11):1970-1982.e7. doi: 10.1016/j.ccell.2024.10.003. Epub 2024 Oct 24. Cancer Cell. 2024. PMID: 39454577 - Research status and development trends of omics in neuroblastoma a bibliometric and visualization analysis.
Han M, Niu H, Duan F, Wang Z, Zhang Z, Ren H. Han M, et al. Front Oncol. 2024 Oct 10;14:1383805. doi: 10.3389/fonc.2024.1383805. eCollection 2024. Front Oncol. 2024. PMID: 39450262 Free PMC article.
References
- Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20. - PubMed
- Matthay KK, et al. Children's Cancer Group Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. New England Journal of Medicine. 1999;341:1165–73. - PubMed
- Schwab M, et al. Chromosome localization in normal human cells and neuroblastomas of a gene related to c-myc. Nature. 1984;308:288–91. - PubMed
- Attiyeh EF, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K08 CA111733/CA/NCI NIH HHS/United States
- R01-CA78454/CA/NCI NIH HHS/United States
- R01 CA078545/CA/NCI NIH HHS/United States
- UL1 RR025774/RR/NCRR NIH HHS/United States
- K08-111733/PHS HHS/United States
- K08 CA111733-04/CA/NCI NIH HHS/United States
- R01-CA87847/CA/NCI NIH HHS/United States
- U10 CA098543/CA/NCI NIH HHS/United States
- R01 CA078545-09/CA/NCI NIH HHS/United States
- R01 CA087847/CA/NCI NIH HHS/United States
- R01 CA124709/CA/NCI NIH HHS/United States
- U10 CA098543-06/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials