Trex1 prevents cell-intrinsic initiation of autoimmunity - PubMed (original) (raw)
Trex1 prevents cell-intrinsic initiation of autoimmunity
Daniel B Stetson et al. Cell. 2008.
Abstract
Detection of nucleic acids and induction of type I interferons (IFNs) are principal elements of antiviral defense but can cause autoimmunity if misregulated. Cytosolic DNA detection activates a potent, cell-intrinsic antiviral response through a poorly defined pathway. In a screen for proteins relevant to this IFN-stimulatory DNA (ISD) response, we identify 3' repair exonuclease 1 (Trex1). Mutations in the human trex1 gene cause Aicardi-Goutieres syndrome (AGS) and chilblain lupus, but the molecular basis of these diseases is unknown. We define Trex1 as an essential negative regulator of the ISD response and delineate the genetic pathway linking Trex1 deficiency to lethal autoimmunity. We show that single-stranded DNA derived from endogenous retroelements accumulates in Trex1-deficient cells, and that Trex1 can metabolize reverse-transcribed DNA. These findings reveal a cell-intrinsic mechanism for initiation of autoimmunity, implicate the ISD pathway as the cause of AGS, and suggest an unanticipated contribution of endogenous retroelements to autoimmunity.
Figures
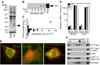
Figure 1. Biochemical identification of Trex1 as an ISD-binding protein
A) Proteins specifically recovered with BrdU-substituted, biotinylated ISD were visualized by silver staining. Arrows indicate the bands identified as Trex1. B) Cells transfected with BrdU-substituted, biotinylated ISD were exposed to ultraviolet light at the indicated times post transfection. Trex1 association with recovered ISD was visualized by western blot. WCE=whole cell extract. C) We examined microarray data (Stetson and Medzhitov, 2006a) of macrophages treated for four hours with ISD and plotted the fold change in expression in log2 format versus the sum of untreated and treated expression values for all 3’->5’ DNA exonucleases in the mouse genome (Shevelev and Hubscher, 2002). Trex1 is indicated in red. D) Bone marrow-derived macrophages of the indicated Trex1 genotype were transfected with ISD. IFN-β and IL-6 mRNA expression four hours post-transfection was measured by quantitative RT-PCR, normalized to HPRT expression within each sample, and compared to untreated controls to calculate the relative expression. E) HeLa cells were transfected with vectors encoding tagged, full-length Trex1 (left panel), Trexl lacking the c-terminal 76 amino acids (middle panel) or YFP fused to the c-terminal 76 amino acids of Trex1 (right panel), and visualized by immunofluorescence microscopy. Images are at 400x magnification. F) 293T cells were transfected with expression vectors for haemagluttinin (HA)-tagged Trexl, HA-Trex1 (1–235), eYFP, or eYFP fused to Trex1 (236–315). Extracts from mechanically disrupted cells were centrifuged through a 10–60% sucrose gradient, and the position of each protein in harvested fractions was determined by western blot for HA (top two panels), eYFP (third and fourth panels), or calnexin (bottom panel).
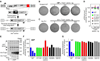
Figure 2. Genetic dissection of the Trex1 knockout phenotype
A) Survival curves for Trex1-deficient mice of the indicated irf3 genotype. Trex1−/− irf3+/+ mice include mice generated both by intercrossing trex1+/− irf3+/− mice as well as by separately intercrossing trex1+/− irf3+/+ mice. These latter mice are also included as “WT” for the crosses in figure 2C and 2D. The number of mice of each genotype is indicated in parentheses, and symbols represent individual mice. B) Body weights of mice of the indicated genotype were measured at 4.5 weeks of age, and compared to the average weight of sex-matched, WT littermates. For these measurements trex1+/− mice were considered WT. C) Survival curves for Trex1-deficient mice of the indicated ifnαr1 genotype. D) Survival curves for Trex1-deficient mice of the indicated rag2 genotype.
Figure 3. Amelioration of autoimmunity through intervention at three phases of disease
A) Paraffin-embedded heart tissue sections from mice of the indicated age and genotype were stained with H&E. The top panels are at 100x and the bottom panels are at 200x magnification. B) IFNβ mRNA levels in hearts of mice of the indicated genotype were determined by quantitative RT-PCR, normalized to HPRT expression within each sample, and compared to WT littermates to calculate the relative IFNβ expression. C) Gene expression analysis of heart tissue from three pairs of Trex1/RAG2 DKO and RAG2 KO littermates, aged 18, 19, and 28 weeks. Data are presented in log2 format and represent means and standard deviations of differential expression in Trex1-deficient hearts compared to wild-type hearts.
Figure 4. The ISD pathway is essential for autoantibodies in Trex1-deficient mice
A) IgG autoantibodies were detected by incubating Trex1/RAG2 DKO heart tissue sections with sera of the indicated genotype and then with fluorescently labeled secondary antibodies to mouse IgG. IgG signal is colored green, and DAPI staining of nuclei is in blue. The images in the left and middle panels are at 200x magnification, and the right panels are 1600x digital zooms of the fields demarcated by the white boxes. B) IgG autoantibodies were detected by western blot against heart protein extracts, prepared from Trex1/RAG2 DKO (KO) or RAG2 KO (WT) mice and used neat or diluted 1:5. C) Heart extracts were prepared as described for Figure 1B and incubated with sera harvested from individual mice of the indicated age and genotype. For each western, the left lane is KO extract and the right lane is WT extract.
Figure 5. Identification of endogenous Trex1 substrates
A) Schematic of the strategy used to tag and amplify DNA fragments purified from heart tissue. B) DNA was purified from pooled hearts of three irf3−/− mice and three trex1−/− irf3−/− mice and PCR-amplified for the indicated number of cycles. The negative control revealed the nonspecific (ns) amplicon, and the positive control amplified a tailed, 58 base DNA oligonucleotide. C) The percentage of unique clones and clones with an endogenous Poly-dT stretch immediately 3’ of the insert are indicated, with the total number of clones per genotype above each bar. D) The relative percentage of each type of DNA fragment within the pool of unique clones. E) Expression levels of 45,037 features from Affymetrix microarray analysis of wild-type heart tissue, compared to the expression levels of the 129 features corresponding to genes with recovered intragenic ssDNA clones, pooled from WT and KO samples. The x-axis is plotted exponentially. F) Assignment and percentages of all recovered clones from WT and KO heart tissue. G) ssDNA fragments that mapped to endogenous retroelements were plotted to scale against a generic, consensus sequence of L1 (top) and LTR (bottom) elements. Sense fragments are above each element, and antisense matches are below. For the fragments that mapped to LTRs, only the 5’ mapping is shown to avoid redundancy.
Figure 6. Trex1 can metabolize endogenous retroelement DNA
A) Schematic of the marked IAP element used to measure IAP retrotransposition efficiency. The L1 element was designed with a similar selectable cassette. B) HeLa cells were transfected with mutant or wild-type marked IAP, alone or together with the indicated amounts of Trex1 expression vector. C) HeLa cells were transfected with the marked L1 element, alone or with the indicated amounts of Trex1 expression vector. D) HeLa cells were transfected with pCDNA3 (which encodes plasmid-based neomycin resistance), alone or with the indicated concentrations of each Trex1 expression vector. E) HEK 293T cells were transfected with the WT IAP, alone or with the indicated concentrations of HA-tagged Trex1. GAG expression and processing 48 hours post transfection was visualized by Western blot. F) Relative IAP retrotransposition efficiency in the presence of the indicated Trex1 expression vectors. For each Trex1 construct, the left bar is 0 ng, the middle bar is 250 ng, and the right bar is 500 ng of expression vector. Data are mean and SEM of triplicate measurements and are representative of five independent experiments. G) Relative L1 retrotransposition efficiency was quantitated in the presence of the indicated mutant forms of Trex1. Data are representative of three independent experiments.
Comment in
- Linking retroelements to autoimmunity.
Bhoj VG, Chen ZJ. Bhoj VG, et al. Cell. 2008 Aug 22;134(4):569-71. doi: 10.1016/j.cell.2008.08.010. Cell. 2008. PMID: 18724930
Similar articles
- Linking retroelements to autoimmunity.
Bhoj VG, Chen ZJ. Bhoj VG, et al. Cell. 2008 Aug 22;134(4):569-71. doi: 10.1016/j.cell.2008.08.010. Cell. 2008. PMID: 18724930 - Cutting Edge: cGAS Is Required for Lethal Autoimmune Disease in the Trex1-Deficient Mouse Model of Aicardi-Goutières Syndrome.
Gray EE, Treuting PM, Woodward JJ, Stetson DB. Gray EE, et al. J Immunol. 2015 Sep 1;195(5):1939-43. doi: 10.4049/jimmunol.1500969. Epub 2015 Jul 29. J Immunol. 2015. PMID: 26223655 Free PMC article. - Aicardi-Goutières syndrome protein TREX1 suppresses L1 and maintains genome integrity through exonuclease-independent ORF1p depletion.
Li P, Du J, Goodier JL, Hou J, Kang J, Kazazian HH Jr, Zhao K, Yu XF. Li P, et al. Nucleic Acids Res. 2017 May 5;45(8):4619-4631. doi: 10.1093/nar/gkx178. Nucleic Acids Res. 2017. PMID: 28334850 Free PMC article. - Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity.
Crow YJ, Rehwinkel J. Crow YJ, et al. Hum Mol Genet. 2009 Oct 15;18(R2):R130-6. doi: 10.1093/hmg/ddp293. Hum Mol Genet. 2009. PMID: 19808788 Free PMC article. Review. - TREX1 As a Potential Therapeutic Target for Autoimmune and Inflammatory Diseases.
Tao SS, Wu GC, Zhang Q, Zhang TP, Leng RX, Pan HF, Ye DQ. Tao SS, et al. Curr Pharm Des. 2019;25(30):3239-3247. doi: 10.2174/1381612825666190902113218. Curr Pharm Des. 2019. PMID: 31475890 Review.
Cited by
- Innate immune sensing of HIV-1 by dendritic cells.
Luban J. Luban J. Cell Host Microbe. 2012 Oct 18;12(4):408-18. doi: 10.1016/j.chom.2012.10.002. Cell Host Microbe. 2012. PMID: 23084911 Free PMC article. Review. - The relationship between defects in DNA repair genes and autoinflammatory diseases.
Kivanc D, Dasdemir S. Kivanc D, et al. Rheumatol Int. 2022 Jan;42(1):1-13. doi: 10.1007/s00296-021-04906-3. Epub 2021 Jun 6. Rheumatol Int. 2022. PMID: 34091703 Review. - DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function.
Yu Q, Katlinskaya YV, Carbone CJ, Zhao B, Katlinski KV, Zheng H, Guha M, Li N, Chen Q, Yang T, Lengner CJ, Greenberg RA, Johnson FB, Fuchs SY. Yu Q, et al. Cell Rep. 2015 May 5;11(5):785-797. doi: 10.1016/j.celrep.2015.03.069. Epub 2015 Apr 23. Cell Rep. 2015. PMID: 25921537 Free PMC article. - Nucleic acid-mediated autoinflammation and autoimmunity-type I interferonopathies.
Lee-Kirsch MA, Günther C, Roers A. Lee-Kirsch MA, et al. J Mol Med (Berl). 2016 Oct;94(10):1081-1084. doi: 10.1007/s00109-016-1467-3. J Mol Med (Berl). 2016. PMID: 27638338 No abstract available. - Caspase activation counteracts interferon signaling after G2 checkpoint abrogation by ATR inhibition in irradiated human cancer cells.
Eek Mariampillai A, Hauge S, Øynebråten I, Rødland GE, Corthay A, Syljuåsen RG. Eek Mariampillai A, et al. Front Oncol. 2022 Oct 28;12:981332. doi: 10.3389/fonc.2022.981332. eCollection 2022. Front Oncol. 2022. PMID: 36387237 Free PMC article.
References
- Alarcon-Riquelme ME. Nucleic acid by-products and chronic inflammation. Nat Genet. 2006;38:866–867. - PubMed
- Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, Demple B, Perrino FW, Lieberman J. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol Cell. 2006;23:133–142. - PubMed
- Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–428. - PubMed
- Colmegna I, Garry RF. Role of endogenous retroviruses in autoimmune diseases. Infect Dis Clin North Am. 2006;20:913–929. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K99 AI072945/AI/NIAID NIH HHS/United States
- R01 AI084914/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- 1K99AI072945-01A1/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases