In vivo reconstitution of autophagy in Saccharomyces cerevisiae - PubMed (original) (raw)
In vivo reconstitution of autophagy in Saccharomyces cerevisiae
Yang Cao et al. J Cell Biol. 2008.
Abstract
Autophagy is a major intracellular degradative pathway that is involved in various human diseases. The role of autophagy, however, is complex; although the process is generally considered to be cytoprotective, it can also contribute to cellular dysfunction and disease progression. Much progress has been made in our understanding of autophagy, aided in large part by the identification of the autophagy-related (ATG) genes. Nonetheless, our understanding of the molecular mechanism remains limited. In this study, we generated a Saccharomyces cerevisiae multiple-knockout strain with 24 ATG genes deleted, and we used it to carry out an in vivo reconstitution of the autophagy pathway. We determined minimum requirements for different aspects of autophagy and studied the initial protein assembly steps at the phagophore assembly site. In vivo reconstitution enables the study of autophagy within the context of the complex regulatory networks that control this process, an analysis that is not possible with an in vitro system.
Figures
Figure 1.
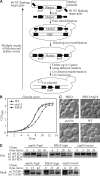
Generation and properties of the MKO strain. (A) Schematic representation of the MKO strategy. The loxP–Cre system allows disruption of up to five genes using different markers, and efficient removal of markers by Cre expression. Multiple rounds of deletion and marker rescue yielded the MKO strain. See Materials and methods for detailed information. (B) Growth curve of the MKO strain compared with wild-type (WT) and atg1Δ strains. WT, atg1Δ, and the MKO strains were grown overnight in YPD and diluted to OD600 = 0.1. Aliquots were removed for OD600 readings every hour for 13 h. (C) Morphology of the MKO strain. WT and atg18Δ strains and the MKO strain with or without a plasmid expressing Atg18 (pATG18(415)) were grown in YPD to mid-log phase and observed by microscopy. The MKO strain had an enlarged vacuole similar to the atg18Δ strain; once transformed with Atg18, the vacuole size became the same as that in the wild type. Bar, 2 μm. (D) The MKO strain expressing Atg6 is not defective for the carboxypeptidase Y, MVB, or alkaline phosphatase pathways, and was assayed by pulse-chase experiments for Prc1, Cps1 (not depicted), and Pho8 processing. The atg6Δ and MKO strains transformed with a plasmid expressing Atg6 (pATG6(414)) or the empty pRS414 vector were subjected to a radioactive label/nonradioactive chase and triple-immunoprecipitated as described in Materials and methods, then analyzed by 8% SDS-PAGE. Carboxypeptidase Y (Prc1) transits through the ER in a precursor (p1) form, acquires its p2 form in the Golgi complex, and, via the late endosome–MVB pathway, is delivered to the vacuole and converted to its mature (m) form. Vacuolar alkaline phosphatase (Pho8) is transported through the ER to the Golgi in a precursor (p) form but bypasses the endosome and is proteolytically activated to its mature (m) form in the vacuole. Atg6 is not required for the alkaline phosphatase pathway; however, for consistency, cells expressing Atg6 were used throughout the experiment.
Figure 2.
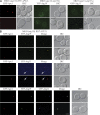
Reconstitution of the cargo recognition and packaging step of the Cvt pathway. (A) Localization of the cargo prApe1 in the MKO (atg15Δ RFP-APE1) strain, and localization of the receptor Atg19 and the adaptor Atg11 in the MKO (ape1Δ atg15Δ) strain. The MKO (atg15Δ RFP-APE1) strain and the MKO (ape1Δ atg15Δ) strain transformed with a plasmid expressing YFP-Atg19 (pYFPATG19(416)) or HA-tagged CFP-Atg11 (pCuHACFPATG11(414)) were grown in selective SMD medium to mid-log phase and observed by fluorescence microscopy. DIC, differential interference contrast. (B) Colocalization of prApe1, Atg19, and Atg11. The MKO (atg15Δ RFP-APE1) strain was transformed with a plasmid expressing YFP-Atg19, HA-tagged CFP-Atg11, or both, grown to mid-log phase and observed by fluorescence microscopy. When Atg19 was coexpressed with prApe1 in the MKO (atg15Δ RFP-APE1) strain, the cargo prApe1 colocalized with the receptor Atg19 (top). When Atg19 was absent, the adaptor Atg11 (arrows) did not colocalize with the cargo (middle). When prApe1, Atg19, and Atg11 were all present, the three proteins colocalized to the same structure (bottom). Bars, 2.5 μm.
Figure 3.
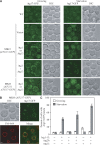
Reconstitution of the initial step of starvation-specific PAS assembly. (A) Localization of Atg17-GFP in the wild-type (WT) and MKO (ATG17-GFP) strains. The WT (ATG17-GFP) strain; the MKO (ATG17-GFP) strain transformed with vector (pRS415), a plasmid expressing Atg1 (pATG1(415)), Atg13 (pATG13(415)), or both Atg1 and Atg13 (pATG1-ATG13(415)); and the MKO (ATG11 ATG17-GFP) strain transformed with a plasmid expressing Atg1 or Atg13 were grown in selective SMD medium to mid-log phase and shifted to SD-N for 2 h for starvation. Samples were taken from both growing (mid-log phase) and starvation conditions for fluorescence microscopy analyses. Atg17-GFP displayed a punctate structure in WT cells in both growing and starvation conditions; some cells had more than one punctum in starvation conditions. In the MKO (ATG17-GFP) strain, Atg17 was cytosolic in both conditions; its localization pattern changed to punctate structures in starvation only when Atg1 and Atg13 were coexpressed. Coexpression of Atg1 and Atg11 or Atg13 and Atg11 could not redistribute Atg17 from the cytosol to punctate structures. Representative images are shown. DIC, differential interference contrast. (B) Perivacuolar localization of Atg17-GFP in the MKO (ATG17-GFP) strain. The MKO (ATG17-GFP) strain transformed with a plasmid expressing both Atg1 and Atg13 (pATG1-ATG13(415)) was grown to mid-log phase, stained with the vacuolar dye FM 4–64 as described in Materials and methods, and shifted to SD-N for 2 h before imaging. When both Atg1 and Atg13 were expressed, Atg17-GFP puncta localized at perivacuolar sites under starvation conditions. Bars, 2.5 μm. (C) The number of cells that contained Atg17-GFP PAS puncta in MKO strains (HCY107, HCY113, and HCY151) with coexpression of various combinations of Atg proteins was quantified under vegetative (open bars) and starvation (closed bars) conditions. Approximately 100–250 cells for each strain were analyzed for scoring the percentage of cells with fluorescent PAS puncta.
Figure 4.
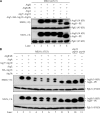
Reconstitution of the Atg12–Atg5 conjugation system. MKO, MKO (ATG3), and atg12Δ cells transformed with various plasmids were grown in selective SMD medium, collected at mid-log phase, and subjected to Western blot analysis using an anti-HA antibody. 0.2 OD600 units of cells were loaded in each lane. Pgk1 was used as a loading control. Plasmids expressing HA-tagged Atg12 (pHA-ATG12(416)); Atg7 and Atg10 (pATG7-ATG10(414)); Atg5 and HA-tagged Atg12 (pATG5-HA-ATG12(416)); Atg5, HA-Atg12, and Atg16 (pATG5-HA-ATG12-ATG16(416)); and Atg8, Atg4, Atg7, and Atg10 (pATG8-ATG4-ATG7-ATG10(414)) were used as indicated. With Atg5, Atg12, Atg7, and Atg10 expressed in the MKO strain, a small amount of Atg12–Atg5 conjugate formed (lane 5). Additional expression of Atg16 improved the conjugation efficiency (lane 6). The presence of Atg8, Atg4, and Atg3 from the Atg8 conjugation system further facilitated the formation and/or enhanced the stability of Atg12–Atg5 (lanes 7 and 8).
Figure 5.
Reconstitution of the Atg8–PE conjugation system. (A) The role of Atg4 and the Atg12–Atg5 conjugation system in Atg8–PE formation. MKO (ATG3) cells transformed with different combinations of plasmids were grown in selective SMD medium, collected at mid-log phase or 2 h after starvation, and then subjected to Western blot analysis using anti-Atg8 antiserum. 0.2 OD600 units of cells were loaded in each lane. Pgk1 was used as a loading control. Plasmids expressing Atg8 (pATG8(414)); Atg8, Atg4, Atg7, and Atg10 (pATG8-ATG4-ATG7-ATG10(414)); Atg5, HA-Atg12, and Atg16 (pATG5-HA-ATG12-ATG16(416)); and Atg8ΔR, Atg7, and Atg10 (pATG8ΔR-ATG7-ATG10(414)) were used as indicated. Atg8–PE was hardly detected in both growing and starvation conditions even when all the known components from the Atg8–PE and Atg12–Atg5 conjugation systems were expressed (lane 4). However, when Atg8ΔR was expressed and Atg4 was absent, a significant amount of Atg8–PE was observed (lane 5), and the amount was further increased when all the components from the Atg12–Atg5 conjugation system were also expressed (lane 6). Note that Atg8–PE migrates aberrantly during SDS-PAGE and runs lower than Atg8. (B) The role of the Atg12–Atg5 conjugation system on Atg8–PE formation. The experimental procedures were the same as in A. Plasmids expressing Atg8ΔR (pATG8ΔR(414)); Atg8ΔR, Atg4, Atg7, and Atg10 (pATG8ΔR-ATG4-ATG7-ATG10(414)); Atg8ΔR, Atg7, and Atg10; Atg5 (pATG5(416)); HA-tagged Atg12 (pHA-ATG12(416)); Atg16 (pATG16(416)); Atg5 and HA-Atg12 (pATG5-HA-ATG12(416)); and Atg5, HA-Atg12, and Atg16 were used as indicated. The atg4Δ atg8Δ strain transformed with the plasmid expressing Atg8ΔR (pATG8ΔR(414)) and the pep4Δ strain were used as controls (lanes 10 and 11). When Atg4 was present, an Atg8–PE band was not detected (compare lanes 3 and 4). Expression of Atg5, Atg12, or Atg16 alone did not improve Atg8–PE formation (compare lanes 5–7 to lane 4). When the Atg12–Atg5 conjugate was formed through the expression of Atg7, Atg10, Atg12, and Atg5, the efficiency of Atg8–PE formation was greatly enhanced (lane 8). Atg16 further facilitated Atg8–PE conjugation and/or enhanced the stability of the conjugate (lane 9).
Similar articles
- New insights into autophagy using a multiple knockout strain.
Cao Y, Klionsky DJ. Cao Y, et al. Autophagy. 2008 Nov;4(8):1073-5. doi: 10.4161/auto.6962. Epub 2008 Nov 10. Autophagy. 2008. PMID: 18971623 Free PMC article. - Monitoring the Formation of Autophagosomal Precursor Structures in Yeast Saccharomyces cerevisiae.
Gómez-Sánchez R, Sánchez-Wandelmer J, Reggiori F. Gómez-Sánchez R, et al. Methods Enzymol. 2017;588:323-365. doi: 10.1016/bs.mie.2016.09.085. Epub 2016 Nov 21. Methods Enzymol. 2017. PMID: 28237109 - Atg27 is a second transmembrane cycling protein.
Yen WL, Klionsky DJ. Yen WL, et al. Autophagy. 2007 May-Jun;3(3):254-6. doi: 10.4161/auto.3823. Epub 2007 May 11. Autophagy. 2007. PMID: 17297289 - Mechanistic Insights into the Role of Atg11 in Selective Autophagy.
Zientara-Rytter K, Subramani S. Zientara-Rytter K, et al. J Mol Biol. 2020 Jan 3;432(1):104-122. doi: 10.1016/j.jmb.2019.06.017. Epub 2019 Jun 22. J Mol Biol. 2020. PMID: 31238043 Free PMC article. Review. - Atg9 trafficking in the yeast Saccharomyces cerevisiae.
Mari M, Reggiori F. Mari M, et al. Autophagy. 2007 Mar-Apr;3(2):145-8. doi: 10.4161/auto.3608. Epub 2007 Mar 21. Autophagy. 2007. PMID: 17204846 Review.
Cited by
- Post-translationally-modified structures in the autophagy machinery: an integrative perspective.
Popelka H, Klionsky DJ. Popelka H, et al. FEBS J. 2015 Sep;282(18):3474-88. doi: 10.1111/febs.13356. Epub 2015 Jul 16. FEBS J. 2015. PMID: 26108642 Free PMC article. Review. - Atg23 and Atg27 act at the early stages of Atg9 trafficking in S. cerevisiae.
Backues SK, Orban DP, Bernard A, Singh K, Cao Y, Klionsky DJ. Backues SK, et al. Traffic. 2015 Feb;16(2):172-90. doi: 10.1111/tra.12240. Epub 2014 Dec 16. Traffic. 2015. PMID: 25385507 Free PMC article. - Autophagy: machinery and regulation.
Yin Z, Pascual C, Klionsky DJ. Yin Z, et al. Microb Cell. 2016 Dec 1;3(12):588-596. doi: 10.15698/mic2016.12.546. Microb Cell. 2016. PMID: 28357331 Free PMC article. Review. - Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay.
Kim M, Sandford E, Gatica D, Qiu Y, Liu X, Zheng Y, Schulman BA, Xu J, Semple I, Ro SH, Kim B, Mavioglu RN, Tolun A, Jipa A, Takats S, Karpati M, Li JZ, Yapici Z, Juhasz G, Lee JH, Klionsky DJ, Burmeister M. Kim M, et al. Elife. 2016 Jan 26;5:e12245. doi: 10.7554/eLife.12245. Elife. 2016. PMID: 26812546 Free PMC article. - Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy.
He C, Baba M, Cao Y, Klionsky DJ. He C, et al. Mol Biol Cell. 2008 Dec;19(12):5506-16. doi: 10.1091/mbc.e08-05-0544. Epub 2008 Oct 1. Mol Biol Cell. 2008. PMID: 18829864 Free PMC article.
References
- Bowers, K., and T.H. Stevens. 2005. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1744:438–454. - PubMed
- Cao, Y., and D.J. Klionsky. 2007. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 17:839–849. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases