miR-19, miR-101 and miR-130 co-regulate ATXN1 levels to potentially modulate SCA1 pathogenesis - PubMed (original) (raw)
. 2008 Oct;11(10):1137-9.
doi: 10.1038/nn.2183. Epub 2008 Aug 31.
Affiliations
- PMID: 18758459
- PMCID: PMC2574629
- DOI: 10.1038/nn.2183
miR-19, miR-101 and miR-130 co-regulate ATXN1 levels to potentially modulate SCA1 pathogenesis
Yoontae Lee et al. Nat Neurosci. 2008 Oct.
Abstract
Spinocerebellar ataxia type 1 is caused by expansion of a translated CAG repeat in ataxin1 (ATXN1). The level of the polyglutamine-expanded protein is one of the factors that contributes to disease severity. Here we found that miR-19, miR-101 and miR-130 co-regulate ataxin1 levels and that their inhibition enhanced the cytotoxicity of polyglutamine-expanded ATXN1 in human cells. We provide a new candidate mechanism for modulating the pathogenesis of neurodegenerative diseases sensitive to protein dosage.
Figures
Figure 1
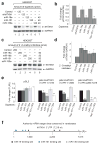
miR-19, miR-101, and miR-130 regulate ATXN1 levels. (a and b) miR-19a, miR-101 and miR-130a decrease ATXN1 level in HEK293T cells. A representative western blot image (a) and mean relative levels of ATXN1 (negative control =1) and standard deviation (s.d.) (b) are presented. (c and d) 2´-O-methyl inhibitors specific for each miRNA increase ATXN1 levels. (e) Luciferase assays using portions of the ATXN1 3´UTR depicted by nucleotide number within the 3´UTR identify the regions regulated by three different miRNAs. (f) Schematic of human ATXN1 3´UTR shows location of the authentic miR-19, miR-101, and miR-130 target sites conserved in vertebrates and verified by mutagenesis in Supplementary Figures 6 and 7. * P≤0.05 and ** P<0.01.
Figure 2
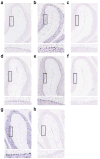
Purkinje cell expression of miR-19, miR-101 and miR-130. In situ hybridization using LNA probes for (a) miR-19b, (b) miR-130a, (c) scrambled (control for miR-19b and miR-130a), (d) miR-101a, (e) miR-101b, (f) scrambled (control for miR-101a and miR-101b), (g) miR-130b, and (e) scrambled (control for miR-130b). Upper panels show miR-19b, miR-101 and miR-130 expression in anterior cerebellar lobules and lower panels are enlarged images of boxed regions to show expression in Purkinje cells.
Figure 3
Inhibition of miRNA-mediated posttranscriptional regulation of polyQ expanded ATXN1 causes more severe cytotoxicity in HEK293T cells. (a) Schematic diagrams of three FLAG-hATXN1[86Q] constructs that either lack the 3´UTR (F86Q), contain a wild type 3´UTR (F86Q-3UTR WT), or contain a 3´UTR harboring mutated miRNA target sites depicted as “x” (F86Q-3UTR Mut). (b and c) A mixture of 2´-O-methyl inhibitors specific for each miRNA (19a+19b+101+130a) increases the level of FLAG-hATXN1[86Q] expressed from F86Q-3UTR WT. A representative western blot image (b) and mean relative levels of FLAG-hATXN1[86Q] (negative control =1) and s.d. (c) are presented. EGFP is a normalization control for transfection efficiency. (d) Lethality is significantly enhanced in cells transfected with the mixture of 2´-O-methyl inhibitors specific for each miRNA and F86Q-3UTR WT compared to cells transfected with 2´-O-methyl inhibitor control and F86Q-3UTR WT. (e and f) Disruption of miRNA target sites in the ATXN1 3´UTR increases FLAG-hATXN1[86Q] level. A representative western blot image (e) and mean relative levels of FLAG-hATXN1[86Q] (F86Q of lane 3 in the western image = 1) and s.d. (f) are presented. (g) Assays of cell viability 48h or 72h after transfection with each vector demonstrate significantly more lethality in cells expressing either F86Q or F86Q-3UTR Mut in comparison to cells expressing F86Q-3UTR WT. * P≤0.05 and ** P<0.01.
Similar articles
- Altered Purkinje cell miRNA expression and SCA1 pathogenesis.
Rodriguez-Lebron E, Liu G, Keiser M, Behlke MA, Davidson BL. Rodriguez-Lebron E, et al. Neurobiol Dis. 2013 Jun;54:456-63. doi: 10.1016/j.nbd.2013.01.019. Epub 2013 Jan 30. Neurobiol Dis. 2013. PMID: 23376683 Free PMC article. - Beyond the glutamine expansion: influence of posttranslational modifications of ataxin-1 in the pathogenesis of spinocerebellar ataxia type 1.
Ju H, Kokubu H, Lim J. Ju H, et al. Mol Neurobiol. 2014 Dec;50(3):866-874. doi: 10.1007/s12035-014-8703-z. Epub 2014 Apr 22. Mol Neurobiol. 2014. PMID: 24752589 Free PMC article. Review. - Purkinje cell ataxin-1 modulates climbing fiber synaptic input in developing and adult mouse cerebellum.
Ebner BA, Ingram MA, Barnes JA, Duvick LA, Frisch JL, Clark HB, Zoghbi HY, Ebner TJ, Orr HT. Ebner BA, et al. J Neurosci. 2013 Mar 27;33(13):5806-20. doi: 10.1523/JNEUROSCI.6311-11.2013. J Neurosci. 2013. PMID: 23536093 Free PMC article. - Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1.
Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY. Lim J, et al. Nature. 2008 Apr 10;452(7188):713-8. doi: 10.1038/nature06731. Epub 2008 Mar 12. Nature. 2008. PMID: 18337722 Free PMC article. - SCA1-phosphorylation, a regulator of Ataxin-1 function and pathogenesis.
Orr HT. Orr HT. Prog Neurobiol. 2012 Dec;99(3):179-85. doi: 10.1016/j.pneurobio.2012.04.003. Epub 2012 Apr 16. Prog Neurobiol. 2012. PMID: 22531670 Free PMC article. Review.
Cited by
- Altered Purkinje cell miRNA expression and SCA1 pathogenesis.
Rodriguez-Lebron E, Liu G, Keiser M, Behlke MA, Davidson BL. Rodriguez-Lebron E, et al. Neurobiol Dis. 2013 Jun;54:456-63. doi: 10.1016/j.nbd.2013.01.019. Epub 2013 Jan 30. Neurobiol Dis. 2013. PMID: 23376683 Free PMC article. - miR-19b promotes tumor growth and metastasis via targeting TP53.
Fan Y, Yin S, Hao Y, Yang J, Zhang H, Sun C, Ma M, Chang Q, Xi JJ. Fan Y, et al. RNA. 2014 Jun;20(6):765-72. doi: 10.1261/rna.043026.113. Epub 2014 Apr 17. RNA. 2014. PMID: 24742936 Free PMC article. - Distinct serum exosomal miRNA profiles detected in acute and asymptomatic dengue infections: A community-based study in Baiyun District, Guangzhou.
Li X, Liao C, Wu J, Yi B, Zha R, Deng Q, Xu J, Guo C, Lu J. Li X, et al. Heliyon. 2024 May 18;10(10):e31546. doi: 10.1016/j.heliyon.2024.e31546. eCollection 2024 May 30. Heliyon. 2024. PMID: 38807894 Free PMC article. - MicroRNA-7 protects against 1-methyl-4-phenylpyridinium-induced cell death by targeting RelA.
Choi DC, Chae YJ, Kabaria S, Chaudhuri AD, Jain MR, Li H, Mouradian MM, Junn E. Choi DC, et al. J Neurosci. 2014 Sep 17;34(38):12725-37. doi: 10.1523/JNEUROSCI.0985-14.2014. J Neurosci. 2014. PMID: 25232110 Free PMC article. - RNA toxicity and foci formation in microsatellite expansion diseases.
Zhang N, Ashizawa T. Zhang N, et al. Curr Opin Genet Dev. 2017 Jun;44:17-29. doi: 10.1016/j.gde.2017.01.005. Epub 2017 Feb 14. Curr Opin Genet Dev. 2017. PMID: 28208060 Free PMC article. Review.
References
- Orr HT, Zoghbi HY. Annu Rev Neurosci. 2007;30:575–621. - PubMed
- Orr HT, et al. Nat Genet. 1993;4:221–6. - PubMed
- Banfi S, et al. Nat Genet. 1994;7:513–20. - PubMed
- Burright EN, et al. Cell. 1995;82:937–48. - PubMed
- Cemal CK, et al. Hum Mol Genet. 2002;11:1075–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NS22920/NS/NINDS NIH HHS/United States
- P30 HD024064-20/HD/NICHD NIH HHS/United States
- R01 NS022920/NS/NINDS NIH HHS/United States
- P30 HD024064/HD/NICHD NIH HHS/United States
- R37 NS022920/NS/NINDS NIH HHS/United States
- NS27699/NS/NINDS NIH HHS/United States
- R37 NS027699/NS/NINDS NIH HHS/United States
- R01 NS027699/NS/NINDS NIH HHS/United States
- R01 NS027699-20/NS/NINDS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- HD24064/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials