The Population Reference Sample, POPRES: a resource for population, disease, and pharmacological genetics research - PubMed (original) (raw)
doi: 10.1016/j.ajhg.2008.08.005. Epub 2008 Aug 28.
Katarzyna Bryc, Karen S King, Amit Indap, Adam R Boyko, John Novembre, Linda P Briley, Yuka Maruyama, Dawn M Waterworth, Gérard Waeber, Peter Vollenweider, Jorge R Oksenberg, Stephen L Hauser, Heide A Stirnadel, Jaspal S Kooner, John C Chambers, Brendan Jones, Vincent Mooser, Carlos D Bustamante, Allen D Roses, Daniel K Burns, Margaret G Ehm, Eric H Lai
Affiliations
- PMID: 18760391
- PMCID: PMC2556436
- DOI: 10.1016/j.ajhg.2008.08.005
The Population Reference Sample, POPRES: a resource for population, disease, and pharmacological genetics research
Matthew R Nelson et al. Am J Hum Genet. 2008 Sep.
Abstract
Technological and scientific advances, stemming in large part from the Human Genome and HapMap projects, have made large-scale, genome-wide investigations feasible and cost effective. These advances have the potential to dramatically impact drug discovery and development by identifying genetic factors that contribute to variation in disease risk as well as drug pharmacokinetics, treatment efficacy, and adverse drug reactions. In spite of the technological advancements, successful application in biomedical research would be limited without access to suitable sample collections. To facilitate exploratory genetics research, we have assembled a DNA resource from a large number of subjects participating in multiple studies throughout the world. This growing resource was initially genotyped with a commercially available genome-wide 500,000 single-nucleotide polymorphism panel. This project includes nearly 6,000 subjects of African-American, East Asian, South Asian, Mexican, and European origin. Seven informative axes of variation identified via principal-component analysis (PCA) of these data confirm the overall integrity of the data and highlight important features of the genetic structure of diverse populations. The potential value of such extensively genotyped collections is illustrated by selection of genetically matched population controls in a genome-wide analysis of abacavir-associated hypersensitivity reaction. We find that matching based on country of origin, identity-by-state distance, and multidimensional PCA do similarly well to control the type I error rate. The genotype and demographic data from this reference sample are freely available through the NCBI database of Genotypes and Phenotypes (dbGaP).
Figures
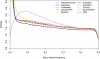
Figure 1
Distribution of Minor-Allele Frequency by Collection Colors and line types for the densities of each collection are shown within the figure.
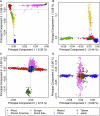
Figure 2
Genetic Structure Illustrated through Scatter Plots of Consecutive Principal Components Subject scores are colored by continental and/or ethnic origin (see legend). East Asian populations are indicated by varying point types. Percent of variation explained by each component is given in parentheses on each axis label.
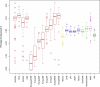
Figure 3
Distribution of Subject-Level Principal Component 5 Scores by Reported Ancestry Each box and whisker indicates the median (heavy line), interquartile range (IQR, box), and minimum and maximum observations (whiskers). Whiskers are truncated at the last observation within 1.5 times the IQR from the edge of the box, with outliers shown individually. Plots for the remaining principal components are available in Figure S2, available online.
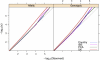
Figure 4
P-Plot Comparing Observed versus Expected Proportion of Associations over a Range of Significance Thresholds Separate lines are presented for each of the four control matching strategies. Results of the allelic exact test are shown on the left and genotypic exact tests on the right. A light gray line corresponds to unity.
Similar articles
- Using population mixtures to optimize the utility of genomic databases: linkage disequilibrium and association study design in India.
Pemberton TJ, Jakobsson M, Conrad DF, Coop G, Wall JD, Pritchard JK, Patel PI, Rosenberg NA. Pemberton TJ, et al. Ann Hum Genet. 2008 Jul;72(Pt 4):535-46. doi: 10.1111/j.1469-1809.2008.00457.x. Epub 2007 May 30. Ann Hum Genet. 2008. PMID: 18513279 Free PMC article. - A panel of 32 AIMs suitable for population stratification correction and global ancestry estimation in Mexican mestizos.
Huerta-Chagoya A, Moreno-Macías H, Fernández-López JC, Ordóñez-Sánchez ML, Rodríguez-Guillén R, Contreras A, Hidalgo-Miranda A, Alfaro-Ruíz LA, Salazar-Fernandez EP, Moreno-Estrada A, Aguilar-Salinas CA, Tusié-Luna T. Huerta-Chagoya A, et al. BMC Genet. 2019 Jan 8;20(1):5. doi: 10.1186/s12863-018-0707-7. BMC Genet. 2019. PMID: 30621578 Free PMC article. - Genome-wide approaches to identify pharmacogenetic contributions to adverse drug reactions.
Nelson MR, Bacanu SA, Mosteller M, Li L, Bowman CE, Roses AD, Lai EH, Ehm MG. Nelson MR, et al. Pharmacogenomics J. 2009 Feb;9(1):23-33. doi: 10.1038/tpj.2008.4. Epub 2008 Feb 26. Pharmacogenomics J. 2009. PMID: 18301416 - The medical and economic roles of pipeline pharmacogenetics: Alzheimer's disease as a model of efficacy and HLA-B(*)5701 as a model of safety.
Roses AD. Roses AD. Neuropsychopharmacology. 2009 Jan;34(1):6-17. doi: 10.1038/npp.2008.153. Epub 2008 Oct 15. Neuropsychopharmacology. 2009. PMID: 18923406 Review. - Genotyping for severe drug hypersensitivity.
Karlin E, Phillips E. Karlin E, et al. Curr Allergy Asthma Rep. 2014 Mar;14(3):418. doi: 10.1007/s11882-013-0418-0. Curr Allergy Asthma Rep. 2014. PMID: 24429903 Free PMC article. Review.
Cited by
- The role of emerging elites in the formation and development of communities after the fall of the Roman Empire.
Tian Y, Koncz I, Defant S, Giostra C, Vyas DN, Sołtysiak A, Pejrani Baricco L, Fetner R, Posth C, Brandt G, Bedini E, Modi A, Lari M, Vai S, Francalacci P, Fernandes R, Steinhof A, Pohl W, Caramelli D, Krause J, Izdebski A, Geary PJ, Veeramah KR. Tian Y, et al. Proc Natl Acad Sci U S A. 2024 Sep 3;121(36):e2317868121. doi: 10.1073/pnas.2317868121. Epub 2024 Aug 19. Proc Natl Acad Sci U S A. 2024. PMID: 39159385 Free PMC article. - Adjusting for principal components can induce spurious associations in genome-wide association studies in admixed populations.
Grinde KE, Browning BL, Reiner AP, Thornton TA, Browning SR. Grinde KE, et al. bioRxiv [Preprint]. 2024 Apr 3:2024.04.02.587682. doi: 10.1101/2024.04.02.587682. bioRxiv. 2024. PMID: 38617337 Free PMC article. Preprint. - Genetic history of East-Central Europe in the first millennium CE.
Stolarek I, Zenczak M, Handschuh L, Juras A, Marcinkowska-Swojak M, Spinek A, Dębski A, Matla M, Kóčka-Krenz H, Piontek J; Polish Archaeogenomics Consortium Team; Figlerowicz M. Stolarek I, et al. Genome Biol. 2023 Jul 24;24(1):173. doi: 10.1186/s13059-023-03013-9. Genome Biol. 2023. PMID: 37488661 Free PMC article. - Inferring human neutral genetic variation from craniodental phenotypes.
Rathmann H, Perretti S, Porcu V, Hanihara T, Scott GR, Irish JD, Reyes-Centeno H, Ghirotto S, Harvati K. Rathmann H, et al. PNAS Nexus. 2023 Jul 3;2(7):pgad217. doi: 10.1093/pnasnexus/pgad217. eCollection 2023 Jul. PNAS Nexus. 2023. PMID: 37457893 Free PMC article. - Demographic inference for spatially heterogeneous populations using long shared haplotypes.
Forien R, Ringbauer H, Coop G. Forien R, et al. bioRxiv [Preprint]. 2023 Jun 13:2023.06.13.544589. doi: 10.1101/2023.06.13.544589. bioRxiv. 2023. PMID: 37398501 Free PMC article. Updated. Preprint.
References
- Jakobsson M., Scholz S.W., Scheet P., Gibbs J.R., VanLiere J.M., Fung H.C., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. - PubMed
- Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. - PubMed
- Manolio T.A., Rodriguez L.L., Brooks L., Abecasis G., Ballinger D., Daly M., Donnelly P., Faraone S.V., Frazer K., Gabriel S. New models of collaboration in genome-wide association studies: The Genetic Association Information Network. Nat. Genet. 2007;39:1045–1051. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM083606-01/GM/NIGMS NIH HHS/United States
- G0700931/MRC_/Medical Research Council/United Kingdom
- G0601966/MRC_/Medical Research Council/United Kingdom
- R01 GM083606/GM/NIGMS NIH HHS/United States
- R01 NS046297/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources