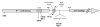Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE Efflux pump system confer different levels of antimicrobial resistance and in vivo fitness - PubMed (original) (raw)
Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE Efflux pump system confer different levels of antimicrobial resistance and in vivo fitness
Douglas M Warner et al. Mol Microbiol. 2008 Oct.
Abstract
The MtrC-MtrD-MtrE efflux pump system confers resistance to macrolide antibiotics and antimicrobial substances of the host innate defence. Clinical isolates with increased resistance to erythromycin and azithromycin frequently harbour mutations in the mtrR structural gene, which encodes a repressor of the mtrCDE operon, or the mtrR promoter region. The MtrC-MtrD-MtrE system is important for gonococcal survival in the murine genital tract, and derepression of the mtrCDE operon via deletion of mtrR confers increased fitness in vivo. Here we compared isogenic strains with naturally occurring mtrR locus mutations for differences in mtrCDE expression and pump-related phenotypes. Mutations upstream of mtrC, including those within the MtrR binding region and a novel mutation that increases mtrC RNA stability conferred the highest levels of derepression as measured by mtrCDE transcription and resistance to antibiotics, progesterone and antimicrobial peptides. In contrast, mutations within the mtrR coding sequence conferred low to intermediate levels of derepression. In vivo, the mtr mutants were more fit than the wild-type strain, the degree to which paralleled in vitro resistance gradients. These studies establish a hierarchy of mtrR locus mutations with regard to regulation of pump efflux, and suggest selection for more derepressed mutants may occur during mixed infections.
Figures
Figure 1. Location of mtr locus mutations used in this study
A schematic of the N. gonorrhoeae mtr locus is shown. The α-helix-encoded region (HTH) of MtrR used for DNA binding is indicated by the hatched pattern. This region is the location of the mutations found in strains DW39, DW45, and MS11. The E202G mutation harbored in strain DW9 is located at the C-terminal end of the MtrR protein, which is hypothesized to be involved in the dimerization of MtrR to itself. The mtrR and mtrCDE transcriptional start sites are indicated by the arrows and are within the intergenic region that contains the MtR binding site and the 13 bp inverted repeat, which is the region where the mutation in strain KH15 is found (Hagman et al., 1995b). The _mtr_120 mutation occurs further upstream of the DNA binding region and is present in strains MS11 and DW120. This mutation has not been described previously, and the G to A change is shown in the detailed DNA sequence.
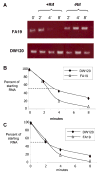
Figure 2. Expression of MtrE by wild type and mtr mutant bacteria
Outer membrane proteins were separated by SDS-PAGE gel electrophoresis and transferred to a PVDF membrane. The 53-kDa MtrE protein was detected by a rabbit MtrE-specific polyclonal antibody described in the Materials and Methods. With the exception of mtrE mutant DW3, protein samples were loaded in ascending order of the levels of antibiotic resistance. The 32-kDa porin protein is constitutively expressed in N. gonorrhoeae and was detected by staining the PVDF membrane with amido black to show equal loading of the samples. The intensity of the MtrE band relative to strain FA19 and after normalization to porin is reported for each strain below the blot. Strain DW11, not described here, carries a frame-shift mutation in the mtrR gene, that is predicted to cause a 52 kD truncated MtrR protein (Warner et al., 2007).
Figure 3. Transcriptional analysis of mtrA, mtrR, mtrC, and rmp
Quantitative RT-PCR was used to assess the fold difference in levels of mRNA compared to the wild type FA19 bacteria. One representative experiment of the three biological replicates tested is shown. There were no significant differences between levels of rmp or mtrA mRNA. mtrR levels were highly down-regulated or non-existent in strains JF1 and KH15 as described previously (Hagman et al., 1995b, Shafer et al., 1995); these values have been omitted to preserve the scale of the figure. Numbers in parentheses denote the fold increase in mtrC levels compared to wild type FA19 bacteria. Levels of mtrC expression were compared using a students t-test to evaluate differences between DW39, DW120, FA19MS11_mtr_, and KH15 gonococci, which exhibit high levels of mtrC expression, and JF1 bacteria, which demonstrate a low-level increase in MIC and mtrC mRNA. ** Denotes p<0.001. The increase in mtrR transcription shown for strain DW120 was 1.7-fold greater than that of strain FA19, but not statistically significant when the averages from three experiments were compared.
Figure 4. Assessment of RNA decay in strain DW120
(A) Equal quantities of RNA from strain FA19 (top) and DW120 (bottom) were reverse transcribed and used in a PCR reaction to determine if there was a difference in the degradation rate of mtrC message. Samples were taken pre (T0) and post rifampicin treatment (+Rif); a second set of cultures to which no rifampicin was added (-Rif) were grown in parallel. (B) RNA samples were also reverse-transcribed and used in a qRT-PCR protocol to quantify amounts of mtrC and rmp RNA from strains FA19 and DW120. These values were used to calculate the RNA half life for mtrC and rmp. The experiment was performed twice and the results were similar.
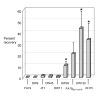
Figure 5. Progesterone resistance
Suspensions of wild type and mutant gonococci were quantitatively cultured on GC agar plates supplemented with progesterone (35 μg/ml) or without progesterone and incubated overnight. Results are expressed as 100 × (# CFU on GC agar with progesterone divided by # CFU on GC alone). The average % recovery calculated from three independent experiments is shown; bars represent the standard error. Asterisks indicate a significant difference between DW39 (p < 0.05) or DW120 and KH15 (p < 0.01) (unpaired t test). The p value for FA19MS11_mtr_ versus wild type FA19 was 0.068.
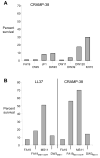
Figure 6. Resistance to antimicrobial peptides
Bacteria were incubated with LL-37, CRAMP, or no peptides and then quantitatively cultured on GC agar. Results are expressed as the percentage of bacteria recovered from wells without peptide, and the graphs shown are representative of one of two or three experiments. Panels A shows the percent recovery of strain FA19 and mtr mutants incubated in CRAMP-38 (4 μg/ml); panel B shows the percent recovery of FA19SmR, FA19MS11mtr, MS11, and DW3MS11 after incubation in LL-37 or CRAMP-38 (16 μg/ml).
Figure 7. Differential in vivo fitness of mtr mutants compared to the wild type strain
Defined ratios of FA19 CmR and mtr mutant bacteria were inoculated into GC broth or estradiol-treated mice to assess the relative fitness of each strain under in vitro and in vivo conditions. The ratio of strains in each inoculum was used in the competitive index (CI) equation as defined in Experimental Procedures. (A) Mixed suspensions were cultured in GC broth (in vitro competition); recovery over the course of growth is expressed as CI. In vivo competition assays between wild type strain FA19CmR and strains (B) DW9, (C) DW39, (D) KH15 and (E) DW120 show different degrees of fitness as measured by CI. Each circle in panels B-E represents the CI from each individual mouse; open circles represent cultures from which no CmR wild type bacteria were recovered. The bars represent the geometric mean of the data, and the dotted line delineates a CI value of 1.0. In cases where a strain was no longer recovered, the limit of detection (4 CFU/100 μl of vaginal wash) was used to calculate the competitive index.
Figure 8. In vivo competition of mtr mutants
Defined ratios of mutant KH15 bacteria and CmR-marked mtr mutants were inoculated into GC broth or estradiol-treated mice to compare the relative fitness under in vitro and in vivo conditions. The ratio of strains in the inoculum was used in the competitive index (CI) equation defined below. (A) Mixed suspensions were cultured in GC broth (in vitro competition); recovery over the course of growth is expressed as CI. The inset shows the optical density of the liquid cultures over time. In vivo competition between strains KH15 and (B) DW9CmR (C) DW45CmR and (D) DW39CmR are shown. In panels B-D, each circle represents the CI value for each individual mouse, and open circles signify mice from which only strain KH15 was recovered, and closed circles represent mice from which both strains were recovered. The CI of 1.0 is denoted by a dashed line, while the geometric mean of each distribution is represented by a bar. In cases where a strain was no longer recovered, the limit of detection (4 CFU/100 μl of vaginal wash) was used to calculate the competitive index. The CI was defined as: (KH15/_mtrR_CmR)output/(KH15/_mtrR_CmR)input, where _mtrR_CmR corresponds to strains DW9CmR, DW45CmR, or DW39CmR.
Similar articles
- Regulation of the MtrC-MtrD-MtrE efflux-pump system modulates the in vivo fitness of Neisseria gonorrhoeae.
Warner DM, Folster JP, Shafer WM, Jerse AE. Warner DM, et al. J Infect Dis. 2007 Dec 15;196(12):1804-12. doi: 10.1086/522964. J Infect Dis. 2007. PMID: 18190261 - Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae.
Veal WL, Nicholas RA, Shafer WM. Veal WL, et al. J Bacteriol. 2002 Oct;184(20):5619-24. doi: 10.1128/JB.184.20.5619-5624.2002. J Bacteriol. 2002. PMID: 12270819 Free PMC article. - A novel mechanism of high-level, broad-spectrum antibiotic resistance caused by a single base pair change in Neisseria gonorrhoeae.
Ohneck EA, Zalucki YM, Johnson PJ, Dhulipala V, Golparian D, Unemo M, Jerse AE, Shafer WM. Ohneck EA, et al. mBio. 2011 Sep 20;2(5):e00187-11. doi: 10.1128/mBio.00187-11. Print 2011. mBio. 2011. PMID: 21933917 Free PMC article. - Azithromycin Resistance through Interspecific Acquisition of an Epistasis-Dependent Efflux Pump Component and Transcriptional Regulator in Neisseria gonorrhoeae.
Wadsworth CB, Arnold BJ, Sater MRA, Grad YH. Wadsworth CB, et al. mBio. 2018 Aug 7;9(4):e01419-18. doi: 10.1128/mBio.01419-18. mBio. 2018. PMID: 30087172 Free PMC article. - Transcriptional regulation of the mtrCDE efflux pump operon: importance for Neisseria gonorrhoeae antimicrobial resistance.
Ayala JC, Balthazar JT, Shafer WM. Ayala JC, et al. Microbiology (Reading). 2022 Aug;168(8):001231. doi: 10.1099/mic.0.001231. Microbiology (Reading). 2022. PMID: 35916832 Free PMC article. Review.
Cited by
- Experimental genital tract infection demonstrates Neisseria gonorrhoeae MtrCDE efflux pump is not required for in vivo human infection and identifies gonococcal colonization bottleneck.
Waltmann A, Balthazar JT, Begum AA, Hua N, Jerse AE, Shafer WM, Hobbs MM, Duncan JA. Waltmann A, et al. PLoS Pathog. 2024 Sep 25;20(9):e1012578. doi: 10.1371/journal.ppat.1012578. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39321205 Free PMC article. Clinical Trial. - Molecular Mechanisms of Bacterial Resistance to Antimicrobial Peptides in the Modern Era: An Updated Review.
Tajer L, Paillart JC, Dib H, Sabatier JM, Fajloun Z, Abi Khattar Z. Tajer L, et al. Microorganisms. 2024 Jun 21;12(7):1259. doi: 10.3390/microorganisms12071259. Microorganisms. 2024. PMID: 39065030 Free PMC article. Review. - The Evaluation of Teleost-Derived Antimicrobial Peptides Against Neisseria gonorrhoeae.
Huang PW, Liou CY, Lee YC, Wei TY, Ho HC, Yang TY, Wang LC. Huang PW, et al. Cureus. 2024 Mar 29;16(3):e57168. doi: 10.7759/cureus.57168. eCollection 2024 Mar. Cureus. 2024. PMID: 38681331 Free PMC article. - Structural analysis of resistance-nodulation cell division transporters.
Klenotic PA, Yu EW. Klenotic PA, et al. Microbiol Mol Biol Rev. 2024 Jun 27;88(2):e0019823. doi: 10.1128/mmbr.00198-23. Epub 2024 Mar 29. Microbiol Mol Biol Rev. 2024. PMID: 38551344 Review. - Rolling the evolutionary dice: Neisseria commensals as proxies for elucidating the underpinnings of antibiotic resistance mechanisms and evolution in human pathogens.
Frost KM, Charron-Smith SL, Cotsonas TC, Dimartino DC, Eisenhart RC, Everingham ET, Holland EC, Imtiaz K, Kornowicz CJ, Lenhard LE, Lynch LH, Moore NP, Phadke K, Reed ML, Smith SR, Ward LL, Wadsworth CB. Frost KM, et al. Microbiol Spectr. 2024 Feb 6;12(2):e0350723. doi: 10.1128/spectrum.03507-23. Epub 2024 Jan 5. Microbiol Spectr. 2024. PMID: 38179941 Free PMC article.
References
- Aas FE, Wolfgang M, Frye S, Dunham S, Lovold C, Koomey M. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol Microbiol. 2002;46:749–760. - PubMed
- Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. - PubMed
- CDC. Increases in fluoroquinolone-resistant Neisseria gonorrhoeae among men who have sex with men--United States, 2003, and revised recommendations for gonorrhea treatment, 2004. MMWR Morb Mortal Wkly Rep. 2004;53:335–338. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 AI021150/AI/NIAID NIH HHS/United States
- U19 AI31496/AI/NIAID NIH HHS/United States
- R01-AI062755/AI/NIAID NIH HHS/United States
- R01-AI42053/AI/NIAID NIH HHS/United States
- U19 AI031496/AI/NIAID NIH HHS/United States
- U19 AI031496-170015/AI/NIAID NIH HHS/United States
- R01 AI042053/AI/NIAID NIH HHS/United States
- R01 AI062755-04/AI/NIAID NIH HHS/United States
- R01 AI062755/AI/NIAID NIH HHS/United States
- R01 AI042053-09/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical