Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses - PubMed (original) (raw)
. 2008 Sep;65(9):996-1006.
doi: 10.1001/archpsyc.65.9.996.
Perciliz L Tan, Ken-ichiro Kubo, Caitlin Engelhard, Koko Ishizuka, Akiharu Kubo, Sachiko Tsukita, Ann E Pulver, Kazunori Nakajima, Nicola G Cascella, Nicholas Katsanis, Akira Sawa
Affiliations
- PMID: 18762586
- PMCID: PMC2727928
- DOI: 10.1001/archpsyc.65.9.996
Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses
Atsushi Kamiya et al. Arch Gen Psychiatry. 2008 Sep.
Abstract
Context: A role for the centrosome has been suggested in the pathology of major mental illnesses, especially schizophrenia (SZ).
Objectives: To show that pericentriolar material 1 protein (PCM1) forms a complex at the centrosome with disrupted-in-schizophrenia 1 (DISC1) and Bardet-Biedl syndrome 4 protein (BBS4), which provides a crucial pathway for cortical development associated with the pathology of SZ. To identify mutations in the PCM1 gene in an SZ population.
Design: Interaction of DISC1, PCM1, and BBS proteins was assessed by immunofluorescent staining and coimmunoprecipitation. Effects of PCM1, DISC1, and BBS on centrosomal functions and corticogenesis in vivo were tested by RNA interference. The PCM1 gene was examined by sequencing 39 exons and flanking splice sites.
Setting: Probands and controls were from the collection of one of us (A.E.P.).
Patients: Thirty-two probands with SZ from families that had excess allele sharing among affected individuals at 8p22 and 219 white controls.
Main outcome measures: Protein interaction and recruitment at the centrosome in cells; neuronal migration in the cerebral cortex; and variant discovery in PCM1 in patients with SZ.
Results: PCM1 forms a complex with DISC1 and BBS4 through discrete binding domains in each protein. DISC1 and BBS4 are required for targeting PCM1 and other cargo proteins, such as ninein, to the centrosome in a synergistic manner. In the developing cerebral cortex, suppression of PCM1 leads to neuronal migration defects, which are phenocopied by the suppression of either DISC1 or BBS4 and are exacerbated by the concomitant suppression of both. Furthermore, a nonsense mutation that segregates with SZ spectrum psychosis was found in 1 family.
Conclusions: Our data further support for the role of centrosomal proteins in cortical development and suggest that perturbation of centrosomal function contributes to the development of mental diseases, including SZ.
Figures
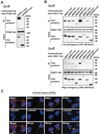
Figure 1. PCM1, DISC1, and BBS proteins interact, localizing with γ-tubulin at the centrosome
(A) PCM1 interacts with DISC1. HA-tagged PCM1 was co-expressed with myc-tagged DISC1 in HEK293 cells. Cell extracts were immunoprecipitated with an anti-HA antibody. Immunoprecipitates were analyzed by Western blotting with an anti-myc antibody (upper panel). The input of each protein is also shown (middle and bottom panels). (B) BBS proteins interact with DISC1 in co-immunopecipitation in HEK293 cells. Myc-tagged BBS1, 2, 4, 5, 6, 7, and 8 all bind to HA-tagged DISC1. Consistent results were observed in both under low stringency (150mM NaCl) washing condition (upper panels) as well as high stringency (500mM NaCl) washing condition (lower panels). The input of each protein is shown in the middle and bottom panels. (C) Localization of BBS1, BBS4, DISC1, and PCM1 in immature cortical neurons at 3 days in vitro (DIV3). Endogenous BBS1, BBS4, and DISC1 (red) are co-localized with γ-tubulin at the centrosome (arrowheads). Endogenous PCM1 (red) mainly occurs just adjacent to γ-tubulin with slight overlap with each other. Blue, nucleus; green, γ-tubulin. Scale bar, 10 µm.
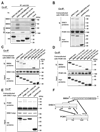
Figure 2. PCM1, DISC1, and BBS4 interact with each other through distinct binding domains
(A) The middle portion of DISC1 (amino acids 349–600) is crucial for DISC1-BBS4 protein interaction. The N-terminal portion (amino acids 1–348) and the C-terminal portion (amino acids 601–854) of DISC1 are important for the DISC1-PCM1 binding. HA-tagged three DISC1 protein fragments [DISC1 (N-348), DISC1 (349–600), and DISC1 (601-C)] were expressed in HEK293 cells for co-immunoprecipitation with an anti-HA antibody. The middle portion of DISC1, but not the N- nor C-terminal DISC1, binds to each of BBS1, 4, and 8, whereas the N- and C-terminal DISC1 bind to PCM1 (upper panels). The inputs of each protein are shown at the right and bottom panels. IB indicates antibodies used for Western blotting. (B) The C-terminus domain of DISC1 for interaction with PCM1 is distinct from the NDEL1 binding domain of DISC1. Deletion of DISC1-NDEL1 binding region [DISC1Δ(802–835)] had no effect on the interaction of DISC1 with PCM1. The inputs are shown in the middle and bottom panels. (C) The second TPR motif of BBS4 is crucial for the BBS4-DISC1 interaction. A series of myc-tagged BBS4 truncation mutants were co-expressed with HA-tagged DISC1 in HEK293 cells for co-immunoprecipitation with an anti-HA antibody. Deletion of the N-terminal region in [BBS4 (1-13-C)] and further deletion of the first TRP motif [BBS4 (2-13-C)] does not affect the BBS4-DISC1 interaction. By contrast, BBS4 mutants with further deletion of the second TRP motif [BBS4 (3-13-C) and BBS4 (4-13-C)] did not bind with DISC1. BBS4 lacking the C-terminal region [BBS4 (N-1-9)] bind to DISC1. The inputs are also shown (middle and bottom panels). (D) The third TPR motif of BBS4 is important for the BBS4-PCM1 interaction. Deletion of the third TPR motif [BBS4 (4-13-C)] dramatically weakened the interaction of myc-tagged BBS4 with HA-tagged PCM1. An anti-HA antibody was used for co-immunoprecipitation. The inputs are shown in the middle and bottom panels. (E) The middle portion of PCM1 is important for DISC1-PCM1 interaction. Myc-tagged DISC1 was co-expressed with HA-tagged PCM1 protein fragments in HEK293 cells for co-immunoprecipitation with an anti-myc antibody. PCM1 (741–1420) has stronger binding affinity to DISC1 than PCM1 (N-740) and PCM1 (1421-C). The inputs of each protein are also shown in middle and bottom panels. (F) Schematic of DISC1, BBS4, and PCM1 interaction shows that these proteins may interact with each other through distinct binding domains.
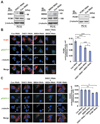
Figure 3. Synergistic effect of DISC1 and BBS4 on recruitment of PCM1 and ninein to the centrosome
(A) Efficient suppression of DISC1, BBS4, and PCM1 by RNAi. RNAi to DISC1, BBS4, and PCM1 suppress 78, 65, and 78% of endogenous DISC1, BBS4, and PCM1 expression, respectively, in PC12 cells (top panels). RNAi to DISC1 or BBS4 does not affect the levels of endogenous PCM1 (middle panels). IB: antibodies used for Western blotting. (B) Suppression of DISC1 and BBS4 reduces accumulation of PCM1 to the centrosome in PC12 cells in a synergistic manner. To quantify the accumulation, immunointensity of PCM1 in the centrosome area (white circle) relative to that in the whole cell region surrounded by green line was quantified. Bars represent averages of each group of cells in three independent and blinded experiments (***P<0.001, ** P<0.01, * P<0.05). Error bars represent SEM. Representative images are shown. Blue, nucleus; red, PCM1; green, pEGFP-F; white, γ-tubulin (also indicated by arrowheads). Scale bar, 10 µm. (C) Accumulation of ninein at the centrosome is disturbed by synergistic application of DISC1 and BBS4 RNAi, or PCM1 RNAi. Although neither application of DISC1 RNAi nor BBS4 RNAi leads to a significant effect on ninein, the synergistic application of both RNAi reduces accumulation of ninein to the centrosome, resembling the phenotype in the presence of RNAi to PCM1. To quantify the accumulation, immunointensity of ninein in the centrosome area (white circle) relative to that in the whole cell region surrounded by the green line was quantified. Bars represent averages of each group of cells in three independent and blinded experiments (* P<0.005, ** P<0.0001). Error bars represent SEM. Representative images of PC12 cells are shown. Blue, nucleus; red, ninein; green, pEGFP-F; white, γ-tubulin (also indicated by arrowheads). Scale bar, 10 µm.
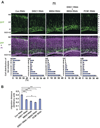
Figure 4. Knockdown of DISC1, BBS4, and PCM1 leads to neuronal migration defects in the developing cerebral cortex
(A) RNAi constructs and GFP expression vectors were electroporated into the ventricular zone (VZ) at E15 and analyzed at P0. In brains with control RNAi (Con RNAi), 40% of GFP-labeled cells exited the VZ, and 25% of GFP-labeled cells completed migration and formed the superficial layers of the cortex that correspond to bins 9 and 10. By contrast, only less than 15% of GFP-positive cells reached the superficial layers in brain slices with DISC1 RNAi, BBS4 RNAi, or PCM1 RNAi, with the majority of GFP-positive cells remaining in the intermediate zone (IZ), subventricular zone (SVZ), and VZ. Green, cells co-transfected with GFP and RNAi constructs; purple, propidium iodide (PI). Scale bar: 100 µm. (B) A migration distance is shown. Silencing of DISC1, BBS4, or PCM1 induces delayed radial migration (** P<0.0001). Silencing of both DISC1 and BBS4 expression leads to a more severe defect compared with that with either DISC1 RNAi or BBS4 RNAi. * P<0.05. Values are mean ± SEM.
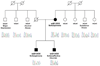
Figure 5. A nonsense mutation in PCM1 in a family with SZ and schizoaffective disorder
Mutation analysis of a Caucasian family JHU37007 shows a heterozygous 4057G→T mutation in exon 24 of PCM1, introducing a premature termination codon (E1353X); genotypes are shown below each individual, as are sequence traces. The psychiatric phenotype (if any) of each family member is also shown.
Similar articles
- The DISC1 Ser704Cys substitution affects centrosomal localization of its binding partner PCM1 in glia in human brain.
Eastwood SL, Walker M, Hyde TM, Kleinman JE, Harrison PJ. Eastwood SL, et al. Hum Mol Genet. 2010 Jun 15;19(12):2487-96. doi: 10.1093/hmg/ddq130. Epub 2010 Apr 1. Hum Mol Genet. 2010. PMID: 20360304 Free PMC article. - The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression.
Kim JC, Badano JL, Sibold S, Esmail MA, Hill J, Hoskins BE, Leitch CC, Venner K, Ansley SJ, Ross AJ, Leroux MR, Katsanis N, Beales PL. Kim JC, et al. Nat Genet. 2004 May;36(5):462-70. doi: 10.1038/ng1352. Epub 2004 Apr 25. Nat Genet. 2004. PMID: 15107855 - A threonine to isoleucine missense mutation in the pericentriolar material 1 gene is strongly associated with schizophrenia.
Datta SR, McQuillin A, Rizig M, Blaveri E, Thirumalai S, Kalsi G, Lawrence J, Bass NJ, Puri V, Choudhury K, Pimm J, Crombie C, Fraser G, Walker N, Curtis D, Zvelebil M, Pereira A, Kandaswamy R, St Clair D, Gurling HM. Datta SR, et al. Mol Psychiatry. 2010 Jun;15(6):615-28. doi: 10.1038/mp.2008.128. Epub 2008 Dec 2. Mol Psychiatry. 2010. PMID: 19048012 - A review of Disrupted-In-Schizophrenia-1 (DISC1): neurodevelopment, cognition, and mental conditions.
Ishizuka K, Paek M, Kamiya A, Sawa A. Ishizuka K, et al. Biol Psychiatry. 2006 Jun 15;59(12):1189-97. doi: 10.1016/j.biopsych.2006.03.065. Biol Psychiatry. 2006. PMID: 16797264 Review. - DISC1-binding proteins in neural development, signalling and schizophrenia.
Bradshaw NJ, Porteous DJ. Bradshaw NJ, et al. Neuropharmacology. 2012 Mar;62(3):1230-41. doi: 10.1016/j.neuropharm.2010.12.027. Epub 2010 Dec 31. Neuropharmacology. 2012. PMID: 21195721 Free PMC article. Review.
Cited by
- Tethering of an E3 ligase by PCM1 regulates the abundance of centrosomal KIAA0586/Talpid3 and promotes ciliogenesis.
Wang L, Lee K, Malonis R, Sanchez I, Dynlacht BD. Wang L, et al. Elife. 2016 May 5;5:e12950. doi: 10.7554/eLife.12950. Elife. 2016. PMID: 27146717 Free PMC article. - Sequencing of DISC1 pathway genes reveals increased burden of rare missense variants in schizophrenia patients from a northern Swedish population.
Moens LN, De Rijk P, Reumers J, Van den Bossche MJ, Glassee W, De Zutter S, Lenaerts AS, Nordin A, Nilsson LG, Medina Castello I, Norrback KF, Goossens D, Van Steen K, Adolfsson R, Del-Favero J. Moens LN, et al. PLoS One. 2011;6(8):e23450. doi: 10.1371/journal.pone.0023450. Epub 2011 Aug 11. PLoS One. 2011. PMID: 21853134 Free PMC article. - Disrupted-in-Schizophrenia-1 Attenuates Amyloid-β Generation and Cognitive Deficits in APP/PS1 Transgenic Mice by Reduction of β-Site APP-Cleaving Enzyme 1 Levels.
Deng QS, Dong XY, Wu H, Wang W, Wang ZT, Zhu JW, Liu CF, Jia WQ, Zhang Y, Schachner M, Ma QH, Xu RX. Deng QS, et al. Neuropsychopharmacology. 2016 Jan;41(2):440-53. doi: 10.1038/npp.2015.164. Epub 2015 Jun 11. Neuropsychopharmacology. 2016. PMID: 26062786 Free PMC article. - Performance of computational methods for the evaluation of pericentriolar material 1 missense variants in CAGI-5.
Monzon AM, Carraro M, Chiricosta L, Reggiani F, Han J, Ozturk K, Wang Y, Miller M, Bromberg Y, Capriotti E, Savojardo C, Babbi G, Martelli PL, Casadio R, Katsonis P, Lichtarge O, Carter H, Kousi M, Katsanis N, Andreoletti G, Moult J, Brenner SE, Ferrari C, Leonardi E, Tosatto SCE. Monzon AM, et al. Hum Mutat. 2019 Sep;40(9):1474-1485. doi: 10.1002/humu.23856. Epub 2019 Aug 17. Hum Mutat. 2019. PMID: 31260570 Free PMC article. - Integrative mechanisms of oriented neuronal migration in the developing brain.
Evsyukova I, Plestant C, Anton ES. Evsyukova I, et al. Annu Rev Cell Dev Biol. 2013;29:299-353. doi: 10.1146/annurev-cellbio-101512-122400. Epub 2013 Aug 7. Annu Rev Cell Dev Biol. 2013. PMID: 23937349 Free PMC article. Review.
References
- Hennah W, Tomppo L, Hiekkalinna T, Palo OM, Kilpinen H, Ekelund J, Tuulio-Henriksson A, Silander K, Partonen T, Paunio T, Terwilliger JD, Lonnqvist J, Peltonen L. Families with the risk allele of DISC1 reveal a link between schizophrenia and another component of the same molecular pathway, NDE1. Hum Mol Genet. 2007;16(5):453–462. - PubMed
- Gurling HM, Critchley H, Datta SR, McQuillin A, Blaveri E, Thirumalai S, Pimm J, Krasucki R, Kalsi G, Quested D, Lawrence J, Bass N, Choudhury K, Puri V, O'Daly O, Curtis D, Blackwood D, Muir W, Malhotra AK, Buchanan RW, Good CD, Frackowiak RS, Dolan RJ. Genetic association and brain morphology studies and the chromosome 8p22 pericentriolar material 1 (PCM1) gene in susceptibility to schizophrenia. Arch Gen Psychiatry. 2006;63(8):844–854. - PMC - PubMed
- Ishizuka K, Paek M, Kamiya A, Sawa A. A review of Disrupted-In-Schizophrenia-1 (DISC1): neurodevelopment, cognition, and mental conditions. Biol Psychiatry. 2006;59(12):1189–1197. - PubMed
- Porteous DJ, Thomson P, Brandon NJ, Millar JK. The genetics and biology of DISC1--an emerging role in psychosis and cognition. Biol Psychiatry. 2006;60(2):123–131. - PubMed
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, Schwab SG, Pulver AE, Faraone SV, Brzustowicz LM, Kaufmann CA, Garver DL, Gurling HM, Lindholm E, Coon H, Moises HW, Byerley W, Shaw SH, Mesen A, Sherrington R, O'Neill FA, Walsh D, Kendler KS, Ekelund J, Paunio T, Lonnqvist J, Peltonen L, O'Donovan MC, Owen MJ, Wildenauer DB, Maier W, Nestadt G, Blouin JL, Antonarakis SE, Mowry BJ, Silverman JM, Crowe RR, Cloninger CR, Tsuang MT, Malaspina D, Harkavy-Friedman JM, Svrakic DM, Bassett AS, Holcomb J, Kalsi G, McQuillin A, Brynjolfson J, Sigmundsson T, Petursson H, Jazin E, Zoega T, Helgason T. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73(1):34–48. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK072301/DK/NIDDK NIH HHS/United States
- R01 MH069853/MH/NIMH NIH HHS/United States
- R01 DK072301/DK/NIDDK NIH HHS/United States
- HD042601/HD/NICHD NIH HHS/United States
- MH-69853/MH/NIMH NIH HHS/United States
- R01 HD042601/HD/NICHD NIH HHS/United States
- R03 MH071473/MH/NIMH NIH HHS/United States
- MH071473/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials