Similar modes of interaction enable Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes - PubMed (original) (raw)
Similar modes of interaction enable Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes
Felix Tritschler et al. Mol Cell Biol. 2008 Nov.
Abstract
Trailer Hitch (Tral or LSm15) and enhancer of decapping-3 (EDC3 or LSm16) are conserved eukaryotic members of the (L)Sm (Sm and Like-Sm) protein family. They have a similar domain organization, characterized by an N-terminal LSm domain and a central FDF motif; however, in Tral, the FDF motif is flanked by regions rich in charged residues, whereas in EDC3 the FDF motif is followed by a YjeF_N domain. We show that in Drosophila cells, Tral and EDC3 specifically interact with the decapping activator DCP1 and the DEAD-box helicase Me31B. Nevertheless, only Tral associates with the translational repressor CUP, whereas EDC3 associates with the decapping enzyme DCP2. Like EDC3, Tral interacts with DCP1 and localizes to mRNA processing bodies (P bodies) via the LSm domain. This domain remains monomeric in solution and adopts a divergent Sm fold that lacks the characteristic N-terminal alpha-helix, as determined by nuclear magnetic resonance analyses. Mutational analysis revealed that the structural integrity of the LSm domain is required for Tral both to interact with DCP1 and CUP and to localize to P-bodies. Furthermore, both Tral and EDC3 interact with the C-terminal RecA-like domain of Me31B through their FDF motifs. Together with previous studies, our results show that Tral and EDC3 are structurally related and use a similar mode to associate with common partners in distinct protein complexes.
Figures
FIG. 1.
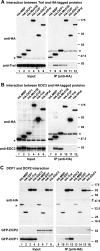
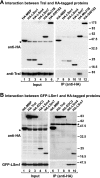
Tral interacts with DCP1, Me31B, and CUP. (A and B) Epitope HA-tagged versions of MBP, DCP1, Me31B, CUP, EDC3 (or Tral), and HA-HPat were transiently expressed in S2 cells, as indicated above the panels. Cell lysates were immunoprecipitated by using a monoclonal anti-HA antibody. Inputs (1.25%) and immunoprecipi- tates (30%) were analyzed by Western blotting using a polyclonal anti-HA antibody. The presence of endogenous Tral (A) or EDC3 (B) in the immunoprecipitates was tested by Western blotting with anti-Tral or anti-EDC3 antibodies. In panel A immunoprecipitations were performed in the presence of RNase A. Asterisks indicate cross-reactivity of the polyclonal anti-HA antibody with an endogenous protein (Input panels) or cross-reactivity with the immunoglobulin heavy chain by the secondary antibody (IP panels). (C) Epitope HA-tagged versions of MBP, GST, Tral, or EDC3 were transiently coexpressed in S2 cells with GFP-DCP1 or GFP-DCP2 as indicated. Cell lysates were immunoprecipitated using a monoclonal anti-HA antibody. Inputs (1.25%) and immunoprecipitates (30%) were analyzed by Western blotting with polyclonal anti-HA and anti-GFP antibodies.
FIG. 2.
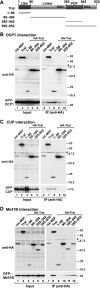
Tral interacts with DCP1, CUP, and Me31B through specific domains. (A) Domain architecture of Tral. Tral homologs contain an LSm domain and an FDF motif. The FDF motif is flanked by sequences rich in glycine and arginine residues (RGG-rich regions). The numbers above the protein outline represent amino acid positions at fragment boundaries for the D. melanogaster protein. The protein domains sufficient for the localization to P bodies and the interaction with DCP1, CUP (1-96), and Me31B (395-542) are indicated. (B to D) HA-tagged Tral or the indicated Tral protein fragments were cotransfected in S2 cells with GFP fusions of DCP1 (B), CUP (C), or Me31B (D) as indicated. Cell lysates were immunoprecipitated using a monoclonal anti-HA antibody. Inputs and immunoprecipitates were analyzed by Western blotting with polyclonal anti-HA and anti-GFP antibodies as described in Fig. 1C.
FIG. 3.
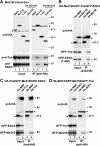
Tral and EDC3 interact with the C-terminal RecA-like domain of Me31B. (A) HA-tagged Me31B or the indicated Me31B protein fragments were cotransfected in S2 cells. Cell lysates were immunoprecipitated with a monoclonal anti-HA antibody. The presence of endogenous Tral or EDC3 in the immunoprecipitates was tested by Western blotting with anti-Tral or anti-EDC3 antibodies as described in Fig. 1A and B. (B to D) S2 cells were cotransfected with mixtures of three plasmids. In panel B the plasmids encoded HA-Me31B, GFP-Tral, and GFP-EDC3 (fragment 1-440). In panel C, the plasmids encoded HA-Tral, GFP-Me31B, and GFP-EDC3. In panel D, the mixture consisted of HA-EDC3, GFP-Me31B, and GFP-Tral. Cell lysates were immunoprecipitated with a monoclonal anti-HA antibody. In all panels, HA-MBP served as a negative control. Inputs and immunoprecipitates were analyzed by Western blotting with polyclonal anti-HA and anti-GFP antibodies as described in Fig. 1C.
FIG. 4.
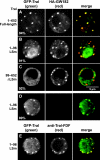
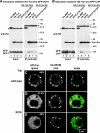
The LSm domain of Tral is necessary and sufficient for P-body localization. (A to E) Confocal fluorescent micrographs of fixed S2 cells expressing GFP-tagged fusions of full-length Tral or the protein fragments indicated on the left. In panels A to C the cells were cotransfected with HA-GW182. In panel E the cells were stained with affinity-purified anti-Tral antibodies (FDF motif). The merged images show the GFP signal in green and the HA or anti-Tral signal in red. The fraction of cells exhibiting a staining identical to that shown in the representative panel was determined by scoring 100 cells in two independent transfections performed per protein. Scale bar, 5 μm.
FIG. 5.
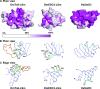
Structure of the LSm domain of D. melanogaster Tral (Tral-LSm). (A) NMR structure of Tral-LSm. (B) NMR structure of the D. melanogaster EDC3 LSm domain (DmEDC3-LSm; PDB-ID: 2rm4) (according to reference 50). (C) Crystal structure of human SmD3 (HsSmD3; PDB-ID: 1d3b-A) (according to reference 20). β-strands belonging to the Sm1 motif are colored in red, and β-strands belonging to the Sm2 motif are colored in yellow (according to references ( and 50). Shown in gray are the N-terminal α-helix in HsSmd3 (that is absent in Tral and EDC3), the extended loop L4 from Tral, and the iβ4 insertion that is unique to EDC3. The GTEx+ motif of Tral is shown in salmon. On the left is a plain view of the open β-barrel with the open side on the top. The center shows the front view, and an edge view is shown on the right.
FIG. 6.
Structure-based alignment of the D. melanogaster Tral LSm domain. (A and B) Superposition of the structures of Tral-LSm (purple), DmEDC3-LSm (lime), and HsSmD3 (gray), represented as tubes. (A) Plain view. (B) Front view (stereo) illustrating the different orientations of loops L3 and L4. (C) Alignment of Tral and EDC3 LSm domains with human SmD3. Secondary structure elements are colored as in Fig. 5. The Sm1 and Sm2 motifs are indicated. Residues from the hydrophobic core that define the Sm1 and Sm2 motifs are shaded blue; conserved glycines are shaded gray. Gray letters indicate amino acids absent in the presented structures. Tral residues mutagenized in the present study, together with residues shown to mediate DCP1 binding in EDC3, are shaded in green. The loop L4 in Tral LSm is boxed in orange, and the GTEx+ motif is shaded in salmon. Black squares indicate acidic residues clustered around Tral residue R21. Abbreviations and accession numbers are as follows: Hs, Homo sapiens (gi:71648673 for RAP55 and gi:74007795 for SmD3); Dm, Drosophila melanogaster (gi:24665977); Gg, Gallus gallus (gi:61098250); Xl, Xenopus laevis (gi:117165637); Dr, Danio rerio (gi:42476222); Ag, Anopheles gambiae (gi:158286595); Ce, Caenorhabditis elegans (gi:17509741); and Sc, Saccharomyces cerevisiae (gi:1066486).
FIG. 7.
The Tral LSm domain is not incorporated into the LSm1-7 ring. (A) HA-tagged versions of MBP, LSm1, LSm4, LSm7, Me31B, and GW182 were transiently expressed in S2 cells. Cell lysates were immunoprecipitated with a monoclonal anti-HA antibody. Inputs and immunoprecipitates were analyzed by Western blotting with a polyclonal anti-HA antibody. The presence of endogenous Tral in the immunoprecipitates was tested by Western blotting with anti-Tral antibodies. (B) HA-tagged versions of MBP, Tral, EDC3, LSm4, and LSm7 were transiently coexpressed in S2 cells with GFP-LSm1. Cell lysates were immunoprecipitated with a monoclonal anti-HA antibody. Inputs and immunoprecipitates were analyzed with polyclonal anti-HA and anti-GFP antibodies.
FIG. 8.
Probing functionally relevant residues in Tral-LSm. (A) Surface representation (plain view) of the structures colored by sequence conservation comparing the seven metazoan species shown in Fig. 6 and Caenorhabditis briggsae. For this figure the yeast orthologs (Scd6p and Edc3p) were excluded because S. cerevisiae Edc3p is highly divergent from metazoa EDC3 (for instance, the linker region between the LSm domain and FDF motif is absent in S. cerevisiae Edc3p, making the alignment with the metazoan proteins difficult). Color ramp by identity: fuchsia (100%) to white (50% or less). (B and C) Tube representation with Cα carbons as spheres in plain view (B) and edge view (C). Loop L4, which is structurally distinct in Tral-LSm and DmEDC3-LSm, is highlighted in orange. Surface residues mutagenized in this and previous studies (50) are drawn as sticks with carbons in green, oxygens in red, and nitrogens in blue. Residues mutagenized in Tral-LSm are located in topologically similar positions to residues in EDC3-LSm that are required for DCP1 binding (Q25, F42, and N44 in EDC3). These residues are on the opposite side from the RNA-binding residues in loops L3 and L5 in canonical (L)Sm proteins (colored cyan on HsSmD3).
FIG. 9.
Effect of Tral-LSm mutations on DCP1 and CUP interaction, as well as on P-body localization. (A and B) HA-tagged MBP, Tral-LSm, or the indicated Tral-LSm mutants were cotransfected in S2 cells with GFP-DCP1 or GFP-CUP, as indicated. Cell lysates were immunoprecipitated with a monoclonal anti-HA antibody and analyzed by Western blotting as described in Fig. 2. Asterisks indicate cross-reactivity of the antibodies as described in Fig. 1. (C to E) Confocal fluorescent micrographs of fixed S2 cells expressing GFP-tagged fusions of full-length Tral wild type or the R21E mutant. Cells were cotransfected with HA-GW182. The merged images show the GFP signal in green and the HA signal in red. The fraction of cells exhibiting a staining identical to that shown in the representative panel was determined by scoring 100 cells in two to three independent transfections performed per protein. Scale bar, 5 μm.
Similar articles
- Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B.
Tritschler F, Braun JE, Eulalio A, Truffault V, Izaurralde E, Weichenrieder O. Tritschler F, et al. Mol Cell. 2009 Mar 13;33(5):661-8. doi: 10.1016/j.molcel.2009.02.014. Mol Cell. 2009. PMID: 19285948 - A divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting.
Tritschler F, Eulalio A, Truffault V, Hartmann MD, Helms S, Schmidt S, Coles M, Izaurralde E, Weichenrieder O. Tritschler F, et al. Mol Cell Biol. 2007 Dec;27(24):8600-11. doi: 10.1128/MCB.01506-07. Epub 2007 Oct 8. Mol Cell Biol. 2007. PMID: 17923697 Free PMC article. - The structural basis of Edc3- and Scd6-mediated activation of the Dcp1:Dcp2 mRNA decapping complex.
Fromm SA, Truffault V, Kamenz J, Braun JE, Hoffmann NA, Izaurralde E, Sprangers R. Fromm SA, et al. EMBO J. 2012 Jan 18;31(2):279-90. doi: 10.1038/emboj.2011.408. Epub 2011 Nov 15. EMBO J. 2012. PMID: 22085934 Free PMC article. - Eukaryotic mRNA Decapping Activation.
Vidya E, Duchaine TF. Vidya E, et al. Front Genet. 2022 Mar 23;13:832547. doi: 10.3389/fgene.2022.832547. eCollection 2022. Front Genet. 2022. PMID: 35401681 Free PMC article. Review. - The role of disordered protein regions in the assembly of decapping complexes and RNP granules.
Jonas S, Izaurralde E. Jonas S, et al. Genes Dev. 2013 Dec 15;27(24):2628-41. doi: 10.1101/gad.227843.113. Genes Dev. 2013. PMID: 24352420 Free PMC article. Review.
Cited by
- CUP promotes deadenylation and inhibits decapping of mRNA targets.
Igreja C, Izaurralde E. Igreja C, et al. Genes Dev. 2011 Sep 15;25(18):1955-67. doi: 10.1101/gad.17136311. Genes Dev. 2011. PMID: 21937713 Free PMC article. - Novel roles of the multi-functional CCR4-NOT complex in post-transcriptional regulation.
Inada T, Makino S. Inada T, et al. Front Genet. 2014 May 20;5:135. doi: 10.3389/fgene.2014.00135. eCollection 2014. Front Genet. 2014. PMID: 24904636 Free PMC article. Review. - Roles of mRNA fate modulators Dhh1 and Pat1 in TNRC6-dependent gene silencing recapitulated in yeast.
Makino S, Mishima Y, Inoue K, Inada T. Makino S, et al. J Biol Chem. 2015 Mar 27;290(13):8331-47. doi: 10.1074/jbc.M114.615088. Epub 2015 Feb 5. J Biol Chem. 2015. PMID: 25657010 Free PMC article. - Molecular basis for GIGYF-Me31B complex assembly in 4EHP-mediated translational repression.
Peter D, Ruscica V, Bawankar P, Weber R, Helms S, Valkov E, Igreja C, Izaurralde E. Peter D, et al. Genes Dev. 2019 Oct 1;33(19-20):1355-1360. doi: 10.1101/gad.329219.119. Epub 2019 Aug 22. Genes Dev. 2019. PMID: 31439631 Free PMC article. - Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation.
Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, Simonelig M, Wahle E. Temme C, et al. RNA. 2010 Jul;16(7):1356-70. doi: 10.1261/rna.2145110. Epub 2010 May 26. RNA. 2010. PMID: 20504953 Free PMC article.
References
- Albrecht, M., and T. Lengauer. 2004. Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett. 56918-26. - PubMed
- Badis, G., C. Saveanu, M. Fromont-Racine, and A. Jacquier. 2004. Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell 155-15. - PubMed
- Barbee, S. A., P. S. Estes, A. M. Cziko, J. Hillebrand, R. A. Luedeman, J. M. Coller, N. Johnson, I. C. Howlett, C. Geng, R. Ueda, A. H. Brand, S. F. Newbury, J. E. Wilhelm, R. B. Levine, A. Nakamura, R. Parker, and M. Ramaswami. 2006. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52997-1009. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous