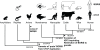The evolution of epigenetic regulators CTCF and BORIS/CTCFL in amniotes - PubMed (original) (raw)
The evolution of epigenetic regulators CTCF and BORIS/CTCFL in amniotes
Timothy A Hore et al. PLoS Genet. 2008.
Abstract
CTCF is an essential, ubiquitously expressed DNA-binding protein responsible for insulator function, nuclear architecture, and transcriptional control within vertebrates. The gene CTCF was proposed to have duplicated in early mammals, giving rise to a paralogue called "brother of regulator of imprinted sites" (BORIS or CTCFL) with DNA binding capabilities similar to CTCF, but testis-specific expression in humans and mice. CTCF and BORIS have opposite regulatory effects on human cancer-testis genes, the anti-apoptotic BAG1 gene, the insulin-like growth factor 2/H19 imprint control region (IGF2/H19 ICR), and show mutually exclusive expression in humans and mice, suggesting that they are antagonistic epigenetic regulators. We discovered orthologues of BORIS in at least two reptilian species and found traces of its sequence in the chicken genome, implying that the duplication giving rise to BORIS occurred much earlier than previously thought. We analysed the expression of CTCF and BORIS in a range of amniotes by conventional and quantitative PCR. BORIS, as well as CTCF, was found widely expressed in monotremes (platypus) and reptiles (bearded dragon), suggesting redundancy or cooperation between these genes in a common amniote ancestor. However, we discovered that BORIS expression was gonad-specific in marsupials (tammar wallaby) and eutherians (cattle), implying that a functional change occurred in BORIS during the early evolution of therian mammals. Since therians show imprinting of IGF2 but other vertebrate taxa do not, we speculate that CTCF and BORIS evolved specialised functions along with the evolution of imprinting at this and other loci, coinciding with the restriction of BORIS expression to the germline and potential antagonism with CTCF.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Gene structure of CTCF and BORIS.
(A) CTCF and BORIS share a similar ZF domain, but different N- and C-terminal domains. (B) All vertebrate CTCF orthologues posses ten exons. Intron-exon boundaries are identical between all CTCF and BORIS orthologues within the ZF domain (grey). Note, genomic coverage of platypus BORIS is incomplete at the 5′ end (*).
Figure 2. Neighbour-joining tree showing relationships between members of the CTCF and BORIS gene family.
Sequence we determined experimentally (blue) and discovered by in silico similarity searches (black) form two distinct clusters with previously annotated CTCF and BORIS orthologues (bold). These clusters are separated from each other by a branch with 100% bootstrap value (thick line). Accession numbers and scientific names for these sequences and species are shown in Table S2.
Figure 3. Human genomic sequence encompassing PCK1-BORIS-RBM38 (top) compared to the orthologous regions in other amniotes.
High similarity over a 100bp window is seen for most exonic sequence (blue) and some untranslated regions (UTR, light blue) or non coding regions (NCR, pink). Despite no similarity to any other region of human BORIS to the chicken PCK1-RBM38 region, the peak labelled with a star is homologous to the first ZF of BORIS.
Figure 4. Expression analysis of CTCF and BORIS.
(A) Conventional RT-PCR of CTCF and BORIS after 35 cycles. Note, cattle brain, platypus ovary and bearded dragon muscle could not be tested due to tissue unavailability or poor RNA quality. (B) BORIS transcript levels relative to the positive control gene GAPDH as quantified by real-time PCR. To assist comparisons between species, we set BORIS expression in the testis to 1 and adjusted all other values within the same species proportionally. As found in humans, expression of BORIS in somatic tissues of cattle and wallaby did not exceed 0.3% of that found in testis, although ovarian expression is just on or above this threshold. In contrast, levels of BORIS expression in platypus (liver and kidney) and bearded dragon (brain, kidney and ovary) are well above this threshold and in some cases match CTCF expression (Figure S2).
Figure 5. Proposed model of CTCF and BORIS evolution in amniotes.
The expression of CTCF and BORIS is indicated (black = expressed, white = not expressed) within various taxa. The ancestral expression pattern of BORIS is wide, including multiple somatic tissues (reptiles and monotremes), but becomes progressively restricted in therian mammals with gonad specific expression in marsupials and cattle (Figure 4) and testis-specific expression in humans and mice ,,. Significant events in the evolution of CTCF and BORIS are marked with respect to the phylogenetic tree.
Similar articles
- The structural complexity of the human BORIS gene in gametogenesis and cancer.
Pugacheva EM, Suzuki T, Pack SD, Kosaka-Suzuki N, Yoon J, Vostrov AA, Barsov E, Strunnikov AV, Morse HC 3rd, Loukinov D, Lobanenkov V. Pugacheva EM, et al. PLoS One. 2010 Nov 8;5(11):e13872. doi: 10.1371/journal.pone.0013872. PLoS One. 2010. PMID: 21079786 Free PMC article. - Early vertebrate origin of CTCFL, a CTCF paralog, revealed by proximity-guided shark genome scaffolding.
Kadota M, Yamaguchi K, Hara Y, Kuraku S. Kadota M, et al. Sci Rep. 2020 Sep 3;10(1):14629. doi: 10.1038/s41598-020-71602-w. Sci Rep. 2020. PMID: 32884037 Free PMC article. - Hypomethylation of the CTCFL/BORIS promoter and aberrant expression during endometrial cancer progression suggests a role as an Epi-driver gene.
Hoivik EA, Kusonmano K, Halle MK, Berg A, Wik E, Werner HM, Petersen K, Oyan AM, Kalland KH, Krakstad C, Trovik J, Widschwendter M, Salvesen HB. Hoivik EA, et al. Oncotarget. 2014 Feb 28;5(4):1052-61. doi: 10.18632/oncotarget.1697. Oncotarget. 2014. PMID: 24658009 Free PMC article. - The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer.
Klenova EM, Morse HC 3rd, Ohlsson R, Lobanenkov VV. Klenova EM, et al. Semin Cancer Biol. 2002 Oct;12(5):399-414. doi: 10.1016/s1044-579x(02)00060-3. Semin Cancer Biol. 2002. PMID: 12191639 Review. - CTCF and BORIS in genome regulation and cancer.
Marshall AD, Bailey CG, Rasko JE. Marshall AD, et al. Curr Opin Genet Dev. 2014 Feb;24:8-15. doi: 10.1016/j.gde.2013.10.011. Epub 2013 Dec 14. Curr Opin Genet Dev. 2014. PMID: 24657531 Review.
Cited by
- The combined action of CTCF and its testis-specific paralog BORIS is essential for spermatogenesis.
Rivero-Hinojosa S, Pugacheva EM, Kang S, Méndez-Catalá CF, Kovalchuk AL, Strunnikov AV, Loukinov D, Lee JT, Lobanenkov VV. Rivero-Hinojosa S, et al. Nat Commun. 2021 Jun 22;12(1):3846. doi: 10.1038/s41467-021-24140-6. Nat Commun. 2021. PMID: 34158481 Free PMC article. - Frequent disruption of chromodomain helicase DNA-binding protein 8 (CHD8) and functionally associated chromatin regulators in prostate cancer.
Damaschke NA, Yang B, Blute ML Jr, Lin CP, Huang W, Jarrard DF. Damaschke NA, et al. Neoplasia. 2014 Dec;16(12):1018-27. doi: 10.1016/j.neo.2014.10.003. Neoplasia. 2014. PMID: 25499215 Free PMC article. - aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity.
Kuraku S, Zmasek CM, Nishimura O, Katoh K. Kuraku S, et al. Nucleic Acids Res. 2013 Jul;41(Web Server issue):W22-8. doi: 10.1093/nar/gkt389. Epub 2013 May 15. Nucleic Acids Res. 2013. PMID: 23677614 Free PMC article. - Differential decay of parent-of-origin-specific genomic sharing in cystic fibrosis-affected sib pairs maps a paternally imprinted locus to 7q34.
Stanke F, Davenport C, Hedtfeld S, Tümmler B. Stanke F, et al. Eur J Hum Genet. 2010 May;18(5):553-9. doi: 10.1038/ejhg.2009.229. Epub 2010 Jan 6. Eur J Hum Genet. 2010. PMID: 20051989 Free PMC article. - The human adenovirus E1B-55K oncoprotein coordinates cell transformation through regulation of DNA-bound host transcription factors.
von Stromberg K, Seddar L, Ip WH, Günther T, Gornott B, Weinert SC, Hüppner M, Bertzbach LD, Dobner T. von Stromberg K, et al. Proc Natl Acad Sci U S A. 2023 Oct 31;120(44):e2310770120. doi: 10.1073/pnas.2310770120. Epub 2023 Oct 26. Proc Natl Acad Sci U S A. 2023. PMID: 37883435 Free PMC article.
References
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, et al. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. - PubMed
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
- Valenzuela L, Kamakaka RT. Chromatin insulators. Annu Rev Genet. 2006;40:107–138. - PubMed
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous