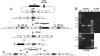A powerful method combining homologous recombination and site-specific recombination for targeted mutagenesis in Drosophila - PubMed (original) (raw)
A powerful method combining homologous recombination and site-specific recombination for targeted mutagenesis in Drosophila
Guanjun Gao et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Gene targeting provides a powerful tool for dissecting gene function. However, repeated targeting of a single locus remains a practice mostly limited to unicellular organisms that afford simple targeting methodologies. We developed an efficient method to repeatedly target a single locus in Drosophila. In this method, which we term "site-specific integrase mediated repeated targeting" (SIRT), an attP attachment site for the phage phiC31 integrase is first targeted to the vicinity of the gene of interest by homologous recombination. All subsequent modifications of that gene are introduced by phiC31-mediated integration of plasmids carrying an attB attachment site and the desired mutation. This highly efficient integration results in a tandem duplication of the target locus, which is then reduced into a single copy carrying the mutation, likely by the efficient "single strand annealing" mechanism, induced with a DNA double-strand break (DSB). We used SIRT to generate a series of six mutations in the Drosophila nbs gene, ranging from single amino acid replacements and small in-frame deletions to complete deletion of the gene. Because all of the components of SIRT are functional in many different organisms, it is readily adaptable to other multicellular organisms.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
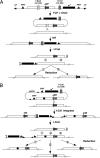
Fig. 1.
A schematic overview of SIRT. (A) Current ends-in targeting scheme followed by reduction for attP placement. At the top is the donor P element. FLP and I-SceI generate a linear extrachromosomal donor molecule. Homologous recombination (HR) integrates the donor into the target locus, creating a tandem duplication. I-CreI cuts between the target copies. Recombination at region 1 leads to an unchanged wild-type copy, whereas recombination at region 2 leads to a copy with the attP site. (B) phiC31-mediated site-specific integration followed by I-CreI-induced reduction. Note that the integrated donor is the entire plasmid carrying vector sequences that include _FRT_s and P ends. The donor plasmid also carries the desired modification (filled circle). Recombination between attP and attB creates attR and attL sites that are no longer the targets for phiC31. I-CreI-induced recombination that occurs at region 1 leads to a target gene carrying the modification and attR. Recombination at region 2 leads to a normal target gene with attR, which can serve as a control locus in subsequent phenotypic analyses. Recombination at region 3 leads to a normal target gene with attL.
Fig. 2.
nbs constructs for SIRT. The genomic regions used to construct the allelic series are shown in the diagram. These regions were cloned into the pTV2 targeting vector giving rise the different M constructs. Coding regions are shown as hatched boxes, with the direction of transcription marked with arrows. Only the first intron of nbs is shown. M0 was used for attP targeting. In M2, serine residues at amino acid (aa) 266 (aa266) and aa268 were mutated to alanine. In M1, M3–5, protein regions deleted were marked with the starting and the ending residues. M40 carried a 40-bp attB site (attB40). The orientation of the nbs inserts are either identical to what is depicted in Fig. 1_B_ or reversed. Frequencies for _vasa-phiC31_-mediated integration by microinjection are calculated as the percentage of fertile crosses with at least one pigmented progeny. The reduction frequencies were calculated as the number of independent reduction lines that retained the desired mutation divided by the total number of independent white-eyed lines established. −FLP, reduction frequencies without an FLP-mediated vector excision step; +FLP, with the FLP step; NA, not applicable; ND, not done.
Fig. 3.
Molecular analyses of products from various SIRT steps. (A–E) Genomic structures for DNA recovered at various steps of SIRT. The distance between the BamHI (B) sites is labeled. The nbs region used as targeting homology is shown as unfilled rectangular boxes. (A) The starting unmodified nbs region. Probe for Southern blotting used in F and G and primer pair (P1 and P2) for long-range PCR in I are indicated. (B) The nbs duplication after ends-in targeting for attP placement. (C) The nbs locus in line nbs434, a reduction product from B that carries attP. (D) The nbs duplication after phiC31-mediated integration. The 5′ and 3′ ends of P element are shown as arrowheads. An nbs mutation is shown as a filled circle. (E) The final SIRT product carrying the mutation and attR. Primer pair (P3 and P4) used in H are shown. (F) Southern blot analysis of M5. Lane m, markers in kb; lane 1, wild-type as in A; lane 2, nbs duplication as in B; lane 3, nbs 434 as in C; lanes 4 and 5, two independent M5 integration lines as in D; lanes 6 and 7, two independent M5 reduction lines as in E. (G) Southern blot analysis of M3. Lane m, markers; lane 1, wild-type; lane 2, nbs 434; lane 3, an M3 integration line as in D; lanes 4 and 5, two independent M3 reduction lines (the original 5.8-kb band is slightly smaller due to the deletion); lane 6, a white-eyed line resulted from NHEJ repair. The 14.5-kb mini-white band harbors a deletion about 8 kb in size (compare lanes 3 and 6). (H) PCR characterization of potential M3 reduction events without the FLP-mediated excision step. The top band was from the wild-type nbs copy, the lower one from the M3 mutant copy. Sample 1 is an M3 integration line as in D. Sample 9 retains both nbs copies. All of the other samples have a precisely reduced nbs locus. (I) Long PCR amplification of reduction events using P1 and P2. Lanes 1 and 2, wild-type control; lane 3, nbs 434 (the product is slightly bigger than wild type due to the presence of V5 and attP); lanes 4–5, two independent final reduction events with the Δ_BRCT1_ mutation (M3); lane 6, a final reduction event with the Δ_BRCT2_ mutation (M5).
Fig. 4.
Using SIRT to precisely delete a gene. (A) Fragment B denotes the gene to be deleted. Fragments A and C are flanking homology used to facilitate recombination. An attP placed between fragments B and C is first targeted to the chromosome. On the injected plasmid donor, an attB is placed between fragments A and B instead. After phiC31-mediated integration, a duplication of fragment B is generated to the left side of the marker gene while a deletion of fragment B is made to the right. During I-CreI-induced reduction, recombination between the two fragment A regions (“region 1”) gives rise to a deletion (left), whereas recombination between the fragment C regions (“region 2”) results in a duplication (right). The positions for primers P1, P2, P5–8 are labeled. (B) PCR analyses on nbs deletion and duplication events. Lane M: markers in kb; lane m, 100-bp ladder. Samples in lanes 1–3 were PCR amplified with P5, P6 (Upper) and P7, P8 (Lower) primer pairs. Samples in lanes 4 and 5 were PCR amplified with P1 and P2. Lane 1 was from a white-eyed fly heterozygous for an nbs duplication. The 2.9-kb product in Lower was from the wild-type nbs locus on the balancer chromosome. Lane 2 was from an nbs deletion heterozygote. Lane 3 was from a homozygous M7 integrant (red-eyed, before reduction). Lane 4 was from a duplication heterozygote, same sample as that in lane 1. No PCR product was expected due to the large size of the fragment. Lane 5 was a deletion heterozygote, same sample as that in lane 2.
Similar articles
- Scarless Modification of the Drosophila Genome Near Any Mapped attP Sites.
Feng S, Mann RS. Feng S, et al. Curr Protoc. 2023 Aug;3(8):e855. doi: 10.1002/cpz1.855. Curr Protoc. 2023. PMID: 37540775 Free PMC article. - Genome manipulations with bacterial recombineering and site-specific integration in Drosophila.
Zhang Y, Schreiner W, Rong YS. Zhang Y, et al. Methods Mol Biol. 2014;1114:11-24. doi: 10.1007/978-1-62703-761-7_2. Methods Mol Biol. 2014. PMID: 24557894 - Long-range targeted manipulation of the Drosophila genome by site-specific integration and recombinational resolution.
Wesolowska N, Rong YS. Wesolowska N, et al. Genetics. 2013 Feb;193(2):411-9. doi: 10.1534/genetics.112.145631. Epub 2012 Nov 12. Genetics. 2013. PMID: 23150601 Free PMC article. - Genome manipulation by homologous recombination in Drosophila.
Bi X, Rong YS. Bi X, et al. Brief Funct Genomic Proteomic. 2003 Jul;2(2):142-6. doi: 10.1093/bfgp/2.2.142. Brief Funct Genomic Proteomic. 2003. PMID: 15239936 Review. - Recombinases and their use in gene activation, gene inactivation, and transgenesis.
Bischof J, Basler K. Bischof J, et al. Methods Mol Biol. 2008;420:175-95. doi: 10.1007/978-1-59745-583-1_10. Methods Mol Biol. 2008. PMID: 18641947 Review.
Cited by
- Initiator elements function to determine the activity state of BX-C enhancers.
Iampietro C, Gummalla M, Mutero A, Karch F, Maeda RK. Iampietro C, et al. PLoS Genet. 2010 Dec 23;6(12):e1001260. doi: 10.1371/journal.pgen.1001260. PLoS Genet. 2010. PMID: 21203501 Free PMC article. - A "mesmer"izing new approach to site-directed mutagenesis in large transformation-ready constructs: Mutagenesis via Serial Small Mismatch Recombineering.
Jacobs JS, Hong X, Eberl DF. Jacobs JS, et al. Fly (Austin). 2011 Apr-Jun;5(2):162-9. doi: 10.4161/fly.5.2.15092. Epub 2011 Apr 1. Fly (Austin). 2011. PMID: 21339708 Free PMC article. - Mre11-Rad50-Nbs complex is required to cap telomeres during Drosophila embryogenesis.
Gao G, Bi X, Chen J, Srikanta D, Rong YS. Gao G, et al. Proc Natl Acad Sci U S A. 2009 Jun 30;106(26):10728-33. doi: 10.1073/pnas.0902707106. Epub 2009 Jun 11. Proc Natl Acad Sci U S A. 2009. PMID: 19520832 Free PMC article. - Fast and efficient Drosophila melanogaster gene knock-ins using MiMIC transposons.
Vilain S, Vanhauwaert R, Maes I, Schoovaerts N, Zhou L, Soukup S, da Cunha R, Lauwers E, Fiers M, Verstreken P. Vilain S, et al. G3 (Bethesda). 2014 Oct 8;4(12):2381-7. doi: 10.1534/g3.114.014803. G3 (Bethesda). 2014. PMID: 25298537 Free PMC article. - FLP recombinase-mediated site-specific recombination in silkworm, Bombyx mori.
Long DP, Zhao AC, Chen XJ, Zhang Y, Lu WJ, Guo Q, Handler AM, Xiang ZH. Long DP, et al. PLoS One. 2012;7(6):e40150. doi: 10.1371/journal.pone.0040150. Epub 2012 Jun 29. PLoS One. 2012. PMID: 22768245 Free PMC article.
References
- Oberstein A, Pare A, Kaplan L, Small S. Site-specific transgenesis by Cre-mediated recombination in Drosophila. Nat Methods. 2005;2:583–585. - PubMed
- Groth AC, Calos MP. Phage integrases: Biology and applications. J Mol Biol. 2004;335:667–678. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous