DNA methylation in development and human disease - PubMed (original) (raw)
Review
DNA methylation in development and human disease
Suhasni Gopalakrishnan et al. Mutat Res. 2008.
Abstract
DNA methylation is a heritable and stable epigenetic mark associated with transcriptional repression. Changes in the patterns and levels of global and regional DNA methylation regulate development and contribute directly to disease states such as cancer. Recent findings provide intriguing insights into the epigenetic crosstalk between DNA methylation, histone modifications, and small interfering RNAs in the control of cell development and carcinogenesis. In this review, we summarize the recent studies in DNA methylation primarily focusing on the interplay between different epigenetic modifications and their potential role in gene silencing in development and disease. Although the molecular mechanisms involved in the epigenetic crosstalk are not fully understood, unraveling their precise regulation is important not only for understanding the underpinnings of cellular development and cancer, but also for the design of clinically relevant and efficient therapeutics using stem cells and anticancer drugs that target tumor initiating cells.
Figures
Fig. 1
The DNA methylation machinery. The mammalian DNA methyltransferase family consists of DNMT1, DNMT3A, DNMT3B, and DNMT3L. DNMT1 is considered the maintenance methyltransferase due to its high activity and preference for hemimethylated DNA during DNA replication. DNMT3A and DNMT3B are de novo methyltransferases. Shown in this figure is a schematic of the DNA methyltransferases with their key functional domains and protein-protein interaction domains indicated. All of the active DNA methyltransferases contain the active site motif IV in the C-terminal region (red box). DNMT1 contains a region required for its interaction with PCNA, which is adjacent to the nuclear localization signal (NLS). The N-terminal region of DNMT1 also contains a cysteine -rich HRX-like region and a lysine-glycine repeat (KG(5)) region. DNMT3A and DNMT3B contain plant homeodomain (PHD) and PWWP domains. These two domains are required for targeting DNMT3A and DNMT3B to pericentromeric heterochromatin and contribute to protein-protein interactions by recognition of histone modifications. Interacting proteins relevant to this review are also listed.
Fig. 2
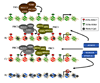
Transcriptional states during differentiation and their associated chromatin modifications. (A) The mammalian trithorax complex (TRX), consisting of the homologs of yeast and Drosophila SET domain containing histone H3K4 methyltransferases MLL1-3 and ASH2L, are responsible for establishing H3K4 trimethylation. H3K4 trimethylation is associated with active transcription represented by the green nucleosomes. (B–C) The polycomb protein complexes PRC1 and PRC2 mediate gene silencing. EZH2 in PRC2 mediates H3K27 trimethylation and also interacts with DNMTs. RING1/2 in the PRC1 complex mediates H2AK119 monoubiquitination. Both H3K27 trimethylation and H2AK119 monoubiquitination are histone marks associated with transcriptional repression (represented by red nucleosomes). Most pluripotency and self-renewal associated genes are marked with H3K4 trimethylation while some differentiation-associated genes are marked bivalently with H3K27 and H3K4 trimethylation. (D) Late differentiation-associated genes are marked by DNA methylation and do not generally contain H3K4 and H3K27 methylation marks (represented by blue nucleosomes). DNA methylation may also play a role in the chromatin states shown in B and C.
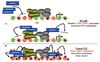
Fig. 3
DNA methyltransferase-polycomb complex interactions in development and cancer. (A) In normal pluripotent cells (ES cells in this case, although tissue-specific stem cells may be similar), genes involved in differentiation are expressed at low levels or are repressed. These genes tend to be marked by both activating (trimethylated H3K4, 3XMe-H3K4) and repressive (trimethylated H3K27, 3XMe-H3K27) marks. A subset of these genes may also be marked by DNA methylation. The regulation of the DNMTs in this state remains unknown. The DNMTs may constitutively associate with polycomb complexes but be rendered inactive or in a low activity state (bottom panel). Alternatively, other chromatin marks or differences in the activity/composition of polycomb complexes prevent DNMT recruitment (top panel). For example, the 3XMe-H3K4 may inhibit DNMT3L-mediated recruitment of de novo DNA methyltransferases. In this bivalent state, genes are able to respond to external developmental cues. (B) In tumor cells or tumor initiating cells (TIC), we propose that normal regulation of the DNMTs is disrupted. This may be due to aberrant recruitment of DNMTs to genes normally repressed by polycomb complexes and/or a change in the activity of bound DNMTs, possibly by post-translational modifications like sumoylation (indicated by the red stars) or a change in the chromatin environment such as acquisition of other repressive histone marks (e.g. trimethylated H3K9). Pro-differentiation genes are then locked in an ‘off’ state due to dense DNA methylation and cells no longer properly respond to differentiation cues and/or they acquire enhanced self-renewal capacity. Once dense DNA methylation is present, the DNMTs may have a role in maintaining PRC1/PRC2 binding.
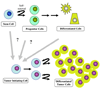
Fig. 4
The cancer stem cell theory. Normal stem cell self-renewal is a tightly regulated process. The resulting progenitor cells are unable to self-renew but can undergo a finite number of divisions and are capable of differentiation. Transformation of a normal stem cell or a dedifferentiated somatic cell by genetic or epigenetic inactivation of tumor suppressor genes or activation of oncogenes can lead to a loss of regulated self-renewal resulting in a cancer stem cell (or tumor initiating cell, TIC), unregulated expansion of the TICs, and aberrant differentiation. It is unknown whether TICs arise from normal stem cells, transiently amplifying progenitors that have gained self-renewal capability, or dedifferentiated somatic cells.
Similar articles
- Epigenetic interplay between histone modifications and DNA methylation in gene silencing.
Vaissière T, Sawan C, Herceg Z. Vaissière T, et al. Mutat Res. 2008 Jul-Aug;659(1-2):40-8. doi: 10.1016/j.mrrev.2008.02.004. Epub 2008 Feb 29. Mutat Res. 2008. PMID: 18407786 Review. - Epigenetic control of embryonic stem cell fate.
Christophersen NS, Helin K. Christophersen NS, et al. J Exp Med. 2010 Oct 25;207(11):2287-95. doi: 10.1084/jem.20101438. J Exp Med. 2010. PMID: 20975044 Free PMC article. Review. - Connections between epigenetic gene silencing and human disease.
Moss TJ, Wallrath LL. Moss TJ, et al. Mutat Res. 2007 May 1;618(1-2):163-74. doi: 10.1016/j.mrfmmm.2006.05.038. Epub 2007 Jan 21. Mutat Res. 2007. PMID: 17306846 Free PMC article. Review. - Epigenetic stem cell signature in cancer.
Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Widschwendter M, et al. Nat Genet. 2007 Feb;39(2):157-8. doi: 10.1038/ng1941. Epub 2006 Dec 31. Nat Genet. 2007. PMID: 17200673 - Epigenetic regulator polycomb group protein complexes control cell fate and cancer.
Kanno R, Janakiraman H, Kanno M. Kanno R, et al. Cancer Sci. 2008 Jun;99(6):1077-84. doi: 10.1111/j.1349-7006.2008.00797.x. Epub 2008 Apr 14. Cancer Sci. 2008. PMID: 18422744 Free PMC article. Retracted. Review.
Cited by
- TERT Promoter Methylation Is Oxygen-Sensitive and Regulates Telomerase Activity.
Dogan F, Forsyth NR. Dogan F, et al. Biomolecules. 2024 Jan 19;14(1):131. doi: 10.3390/biom14010131. Biomolecules. 2024. PMID: 38275760 Free PMC article. - HOXB13, a target of DNMT3B, is methylated at an upstream CpG island, and functions as a tumor suppressor in primary colorectal tumors.
Ghoshal K, Motiwala T, Claus R, Yan P, Kutay H, Datta J, Majumder S, Bai S, Majumder A, Huang T, Plass C, Jacob ST. Ghoshal K, et al. PLoS One. 2010 Apr 29;5(4):e10338. doi: 10.1371/journal.pone.0010338. PLoS One. 2010. PMID: 20454457 Free PMC article. Retracted. - Insulin but not glucagon gene is silenced in human pancreas-derived mesenchymal stem cells.
Wilson LM, Wong SH, Yu N, Geras-Raaka E, Raaka BM, Gershengorn MC. Wilson LM, et al. Stem Cells. 2009 Nov;27(11):2703-11. doi: 10.1002/stem.229. Stem Cells. 2009. PMID: 19785038 Free PMC article. - Genome-wide alteration of 5-hydroxymethylcytosine in a mouse model of fragile X-associated tremor/ataxia syndrome.
Yao B, Lin L, Street RC, Zalewski ZA, Galloway JN, Wu H, Nelson DL, Jin P. Yao B, et al. Hum Mol Genet. 2014 Feb 15;23(4):1095-107. doi: 10.1093/hmg/ddt504. Epub 2013 Oct 9. Hum Mol Genet. 2014. PMID: 24108107 Free PMC article. - On the thermodynamics of DNA methylation process.
Sanchez R, Mackenzie SA. Sanchez R, et al. Sci Rep. 2023 Jun 1;13(1):8914. doi: 10.1038/s41598-023-35166-9. Sci Rep. 2023. PMID: 37264042 Free PMC article.
References
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. - PubMed
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. - PubMed
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. - PubMed
- Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279:27816–27823. - PubMed
- Watanabe D, Suetake I, Tada T, Tajima S. Stage- and cell-specific expression of Dnmt3a and Dnmt3b during embryogenesis. Mech Dev. 2002;118:187–190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01CA116028/CA/NCI NIH HHS/United States
- R01 CA116028/CA/NCI NIH HHS/United States
- R01 CA114229/CA/NCI NIH HHS/United States
- R01 CA114229-04/CA/NCI NIH HHS/United States
- F32CA132327/CA/NCI NIH HHS/United States
- R01 CA116028-03/CA/NCI NIH HHS/United States
- R01CA114229/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases