Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc - PubMed (original) (raw)
Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc
Zihao Wang et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Protein GlcNAcylation serves as a nutrient/stress sensor to modulate the functions of many nuclear and cytoplasmic proteins. O-GlcNAc cycles on serine or threonine residues like phosphorylation, is nearly as abundant, and functions, at least partially, via its interplay with phosphorylation. Here, we describe changes in site-specific phosphorylation dynamics in response to globally elevated GlcNAcylation. By combining sequential phospho-residue enrichment, iTRAQ labeling, and high throughput mass spectrometric analyses, phosphorylation dynamics on 711 phosphopeptides were quantified. Based upon their insensitivity to phosphatase inhibition, we conclude that approximately 48% of these phosphorylation sites were not actively cycling in the conditions of the study. However, increased GlcNAcylation influenced phosphate stoichiometry at most of the sites that did appear to be actively cycling. Elevated GlcNAcylation resulted in lower phosphorylation at 280 sites and caused increased phosphorylation at 148 sites. Thus, the cross-talk or interplay between these two abundant posttranslational modifications is extensive, and may arises both by steric competition for occupancy at the same or proximal sites and by each modification regulating the other's enzymatic machinery. The phosphoproteome dynamics presented by this large set of quantitative data not only delineates the complex interplay between phosphorylation and GlcNAcyation, but also provides insights for more focused investigations of specific roles of O-GlcNAc in regulating protein functions and signaling pathways.
Conflict of interest statement
Conflict of interest statement: G.W.H. receives a share of royalty received by the university on sales of the CTD 110.6 antibody. Terms of this arrangement are managed by Johns Hopkins University School of Medicine.
Figures
Fig. 1.
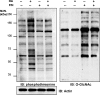
Effects of elevated GlcNAcylation and phosphorylation on each other. NIH 3T3 cells were untreated and treated with okadaic acid (225 nM), PUGNAc (100 μM), and 20 μM NAG-thiazoline as indicated for 2.5 h before harvest. Cell lysates (20 μg for each group) were resolved in SDS/PAGE, transferred, and blotted with phosphothreonine or _O_-GlcNAc antibody. The same membrane was stripped and blotted with anti-actin for loading control. OA, okadaic acid; P/N, combined treatment with PUGNAc and NAG-thiazoline.
Fig. 2.
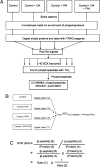
Quantitative proteomics approach used to delineate global interplay between phosphorylation and GlcNAcylation. (A) Flow chart for phosphosite detection and site-specific quantitation. (B) Flow chart for protein expression level quantitation using iTRAQ. (C) Formula used to calculate RORs.
Fig. 3.
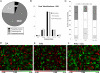
Summary of large-scale identification and quantitation. (A) Distribution of detected phosphoserine, threonine, and tyrosine sites. (B) Total number of identified proteins and distribution of protein expression level dynamics. (C) Distribution of phosphorylation ROR dynamics. (D–F) Log2 ratios of site-specific phosphorylation RORs under indicated conditions. Positions at the heat maps correspond to the same phosphorylation sites.
Fig. 4.
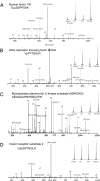
Detection and quantitation of phosphopeptides. Selected samples originating from nuclear factor 1/B (A), DNA replication licensing factor MCM4 (B), myristoylated alanine-rich C-kinase substrate (C), and insulin receptor substrate 2 (D). (Insets) Indicate the intensities of iTRAQ reporter ions.
Fig. 5.
DNA/RNA-binding proteins showed different patterns of phosphorylation dynamics after treatment of _O_-GlcNAcase inhibitors compared with cytosketetal and cytoskeleton-binding proteins.
Similar articles
- Dynamic interplay between O-linked N-acetylglucosaminylation and glycogen synthase kinase-3-dependent phosphorylation.
Wang Z, Pandey A, Hart GW. Wang Z, et al. Mol Cell Proteomics. 2007 Aug;6(8):1365-79. doi: 10.1074/mcp.M600453-MCP200. Epub 2007 May 16. Mol Cell Proteomics. 2007. PMID: 17507370 - Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity.
Copeland RJ, Bullen JW, Hart GW. Copeland RJ, et al. Am J Physiol Endocrinol Metab. 2008 Jul;295(1):E17-28. doi: 10.1152/ajpendo.90281.2008. Epub 2008 Apr 29. Am J Physiol Endocrinol Metab. 2008. PMID: 18445751 Free PMC article. Review. - Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides.
Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. Chalkley RJ, et al. Proc Natl Acad Sci U S A. 2009 Jun 2;106(22):8894-9. doi: 10.1073/pnas.0900288106. Epub 2009 May 19. Proc Natl Acad Sci U S A. 2009. PMID: 19458039 Free PMC article. - Characterization and identification of protein O-GlcNAcylation sites with substrate specificity.
Wu HY, Lu CT, Kao HJ, Chen YJ, Chen YJ, Lee TY. Wu HY, et al. BMC Bioinformatics. 2014;15 Suppl 16(Suppl 16):S1. doi: 10.1186/1471-2105-15-S16-S1. Epub 2014 Dec 8. BMC Bioinformatics. 2014. PMID: 25521204 Free PMC article. - Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease.
Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Hart GW, et al. Annu Rev Biochem. 2011;80:825-58. doi: 10.1146/annurev-biochem-060608-102511. Annu Rev Biochem. 2011. PMID: 21391816 Free PMC article. Review.
Cited by
- Investigation of in vitro histone H3 glycosylation using H3 tail peptides.
Merx J, Hintzen JCJ, Proietti G, Elferink H, Wang Y, Porzberg MRB, Sondag D, Bilgin N, Park J, Mecinović J, Boltje TJ. Merx J, et al. Sci Rep. 2022 Nov 10;12(1):19251. doi: 10.1038/s41598-022-21883-0. Sci Rep. 2022. PMID: 36357422 Free PMC article. - Overview: the maturing of proteomics in cardiovascular research.
Van Eyk JE. Van Eyk JE. Circ Res. 2011 Feb 18;108(4):490-8. doi: 10.1161/CIRCRESAHA.110.226894. Circ Res. 2011. PMID: 21335431 Free PMC article. Review. - Regulation of autophagy by protein post-translational modification.
Wani WY, Boyer-Guittaut M, Dodson M, Chatham J, Darley-Usmar V, Zhang J. Wani WY, et al. Lab Invest. 2015 Jan;95(1):14-25. doi: 10.1038/labinvest.2014.131. Epub 2014 Nov 3. Lab Invest. 2015. PMID: 25365205 Free PMC article. Review. - The emerging link between O-GlcNAcylation and neurological disorders.
Ma X, Li H, He Y, Hao J. Ma X, et al. Cell Mol Life Sci. 2017 Oct;74(20):3667-3686. doi: 10.1007/s00018-017-2542-9. Epub 2017 May 22. Cell Mol Life Sci. 2017. PMID: 28534084 Free PMC article. Review. - Mass spectrometry in the analysis of N-linked and O-linked glycans.
North SJ, Hitchen PG, Haslam SM, Dell A. North SJ, et al. Curr Opin Struct Biol. 2009 Oct;19(5):498-506. doi: 10.1016/j.sbi.2009.05.005. Epub 2009 Jul 3. Curr Opin Struct Biol. 2009. PMID: 19577919 Free PMC article. Review.
References
- Hart G, Housley M, Slawson C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. - PubMed
- Slawson C, Housley M, Hart G. O-GlcNAc cycling: How a single sugar post-translational modification is changing the way we think about signaling networks. J Cell Biochem. 2006;97:71–83. - PubMed
- Zachara N, Hart G. O-GlcNAc a sensor of cellular state: The role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim Biophys Acta. 2004;1673:13–28. - PubMed
- Kreppel L, Hart G. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J Biol Chem. 1999;274:32015–32022. - PubMed
- Gao Y, Wells L, Comer F, Parker G, Hart G. Dynamic O-glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a neutral, cytosolic β-N-acetylglucosaminidase from human brain. J Biol Chem. 2000;276:9838–9845. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases