Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription - PubMed (original) (raw)
Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription
Gary Shaw et al. Structure. 2008.
Abstract
RapA, as abundant as sigma70 in the cell, is an RNA polymerase (RNAP)-associated Swi2/Snf2 protein with ATPase activity. It stimulates RNAP recycling during transcription. We report a structure of RapA that is also a full-length structure for the entire Swi2/Snf2 family. RapA contains seven domains, two of which exhibit novel protein folds. Our model of RapA in complex with ATP and double-stranded DNA (dsDNA) suggests that RapA may bind to and translocate on dsDNA. Our kinetic template-switching assay shows that RapA facilitates the release of sequestered RNAP from a posttranscrption/posttermination complex for transcription reinitiation. Our in vitro competition experiment indicates that RapA binds to core RNAP only but is readily displaceable by sigma70. RapA is likely another general transcription factor, the structure of which provides a framework for future studies of this bacterial Swi2/Snf2 protein and its important roles in RNAP recycling during transcription.
Figures
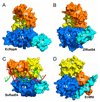
Figure 1. Overall structure of E. coli RapA
On the top, the domain organization of RapA is shown with boundaries (residue numbers in the sequence). In the middle, the domain arrangement is shown schematically by two views of the molecule related to each other by a 180° rotation around the vertical axis. On the left, right and bottom, the folds of individual domains and a cluster of four domains (1A, 1B, 2A, and 2B, which form the ATPase core of the enzyme) are illustrated. Both the orientation and the scale of the individual domains and the ATPase core are optimized for best viewing. The Ntd (N-terminal domain) contains two sub-domains, NtdA and NtdB, which share the same fold. The Linker3 is illustrated as a stick model (C, grey; N, blue; O, red) outlined with the composite annealed omit map (green net, 2Fo - Fc, contoured at 1.0 σ). The Spacer and the Ctd (C-terminal domain) exhibit novel folds. The sulfate ion bound to the ATPase core is shown as a sphere model. The color scheme used here (Ntd in red, Linker1 in black, 1A in blue, 1B in cyan, Linker2 in green, 2A in yellow, 2B in orange, Linker3 in magenta, Spacer in green and Ctd in grey) is also used in other figures.
Figure 2. Structure and sequence characteristics of RapA
(A) Stereoview shows the Cα trace of RapA. Every twentieth Cα position is labeled with the tenth Cα position between every two labels indicated with a sphere. The domain structure of the protein is color coded as done in Figure 1. The sulfate ion is shown as a stick model in black. (B) Sequence of RapA with secondary structure elements indicated above. One N-terminal residue and six C-terminal residues were not observed. Underlined are the six functional motifs located in the RecA-like domains 1A and 2A of the ATPase core.
Figure 3. RecA-like architecture and the six conserved functional motifs
(A–D) The RacA-like architecture of DExx box helicase PcrA in complex with DNA and an ATP analog (PDB entry 2PJR), ZfRad54 in complex with a sulfate ion (PDB entry 1Z3I), SsRad54 in complex with dsDNA (PDB entry 1Z63), and E. coli RapA in complex with a sulfate ion are illustrated as Cα traces on the basis of the superposition of domain 1A. Domain 1A is shown in blue and 2A in yellow. Indicated in red are the six functional motifs, the sequences of which are shown in panel E. The ATP and SO4 are shown as stick models while DNA molecules as tubes. (E) Structure-based sequence alignment for the six functional motifs in PcrA•ATP•DNA, ZfRad54•SO4, SsRad54•DNA and RapA•SO4. Amino acid residues highlighted in red are also shown in red in panels A–D. The functional motifs are labeled in panel D only. The roles (interactions) of the six functional motifs are predicted on the basis of helicase structures (Caruthers and McKay, 2002).
Figure 4. The ATPase core of Swi2/Snf2 proteins RapA and Rad54
(A–D) Domains 1A, 1B, 2A and 2B in the ATPase core of E. coli RapA, ZfRad54 (PDB entry 1Z3I), SsRad54•DNA (PDB entry 1Z63), and E. coli Mfd (PDB entry 2EYQ) are illustrated as surface representation. The dsDNA in panel c is shown as a tube-and-stick model.
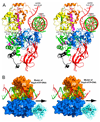
Figure 5. Hypothetical model of the RapA•ATP•dsDNA complex
(A) The ATPase core of RapA (ribbon diagram in the same color scheme defined in Figure 1) is shown with superimposed dsDNA (tube-and-stick model) from the SsRad54•DNA structure (PDB entry 1Z63), and the ATP (stick model) and a portion of ssDNA (tube-and-stick model) from the PcrA•ATP•DNA structure (PDB entry 3PJR). The SsRad54 and the PcrA are not shown for clarity. (B) The RapA•ATP•dsDNA model suggests a role of domain 2B in ATP-binding-associated advancement of dsDNA for Swi2/Snf2 proteins. In this view, the ATP molecule is not visible. The white arrow suggests the transporting direction of DNA; the black arrow suggests the translocation direction of the ATPase core along the minor groove of dsDNA. For clarity, only the ATPase core is shown instead of the full-length structure.
Figure 6. RapA facilitates the release of RNAP in an in vitro DNA template-switch transcription assay
The reaction was started (as time 0) by the addition of NTPs into a preincubated complex of RNAP and a plasmid DNA containing pTac (the first template) either in the presence (+) or absence (−) of RapA; and at various times as indicated, an equal molar amount of another plasmid DNA containing λ_PL_ (as a second template) was added, followed by 1 hr incubation for the reaction to complete. The transcripts for pTac and λ_PL_ are indicated. The RNAI transcript from a relatively weak promoter present in both plasmids is also indicated.
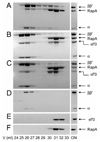
Figure 7. Gel filtration analyses: RapA and σ70 compete for binding to CORE in vitro
The loaded protein mixture is indicated with the word ON. The positions for subunits of CORE, RapA and σ70 are indicated. The concentration of each purified protein used in the representative set of experiments was 1 µM. (A) RapA + CORE. (B) (RapA + σ70) + CORE: RapA and σ70 were premixed on ice followed by incubation with CORE at room temperature for 15 min. (C) [RapA+ CORE] + σ70: RapA and CORE were premixed and incubated at room temperature for 15 min, followed by the addition of σ70 and incubation at room temperature for another 15 min. (D–F) Controls of the experiment for CORE, σ70, and RapA, respectively.
Comment in
- RapA: completing the transcription cycle?
Nechaev S, Severinov K. Nechaev S, et al. Structure. 2008 Sep 10;16(9):1294-5. doi: 10.1016/j.str.2008.08.001. Structure. 2008. PMID: 18786392 No abstract available.
Similar articles
- Structure and function of RapA: a bacterial Swi2/Snf2 protein required for RNA polymerase recycling in transcription.
Jin DJ, Zhou YN, Shaw G, Ji X. Jin DJ, et al. Biochim Biophys Acta. 2011 Sep;1809(9):470-5. doi: 10.1016/j.bbagrm.2011.03.003. Epub 2011 Mar 16. Biochim Biophys Acta. 2011. PMID: 21419241 Free PMC article. Review. - Structural basis for activation of Swi2/Snf2 ATPase RapA by RNA polymerase.
Shi W, Zhou W, Chen M, Yang Y, Hu Y, Liu B. Shi W, et al. Nucleic Acids Res. 2021 Oct 11;49(18):10707-10716. doi: 10.1093/nar/gkab744. Nucleic Acids Res. 2021. PMID: 34428297 Free PMC article. - Allosteric Activation of Bacterial Swi2/Snf2 (Switch/Sucrose Non-fermentable) Protein RapA by RNA Polymerase: BIOCHEMICAL AND STRUCTURAL STUDIES.
Kakar S, Fang X, Lubkowska L, Zhou YN, Shaw GX, Wang YX, Jin DJ, Kashlev M, Ji X. Kakar S, et al. J Biol Chem. 2015 Sep 25;290(39):23656-69. doi: 10.1074/jbc.M114.618801. Epub 2015 Aug 13. J Biol Chem. 2015. PMID: 26272746 Free PMC article. - Recycling of bacterial RNA polymerase by the Swi2/Snf2 ATPase RapA.
Inlow K, Tenenbaum D, Friedman LJ, Kondev J, Gelles J. Inlow K, et al. Proc Natl Acad Sci U S A. 2023 Jul 11;120(28):e2303849120. doi: 10.1073/pnas.2303849120. Epub 2023 Jul 5. Proc Natl Acad Sci U S A. 2023. PMID: 37406096 Free PMC article. - σ70 and PhoB activator: getting a better grip.
Canals A, Blanco AG, Coll M. Canals A, et al. Transcription. 2012 Jul-Aug;3(4):160-4. doi: 10.4161/trns.20444. Epub 2012 Jul 1. Transcription. 2012. PMID: 22771992 Free PMC article. Review.
Cited by
- RNA polymerase-promoter interactions determining different stability of the Escherichia coli and Thermus aquaticus transcription initiation complexes.
Mekler V, Minakhin L, Kuznedelov K, Mukhamedyarov D, Severinov K. Mekler V, et al. Nucleic Acids Res. 2012 Dec;40(22):11352-62. doi: 10.1093/nar/gks973. Epub 2012 Oct 18. Nucleic Acids Res. 2012. PMID: 23087380 Free PMC article. - Structural basis for recognition and remodeling of the TBP:DNA:NC2 complex by Mot1.
Butryn A, Schuller JM, Stoehr G, Runge-Wollmann P, Förster F, Auble DT, Hopfner KP. Butryn A, et al. Elife. 2015 Aug 10;4:e07432. doi: 10.7554/eLife.07432. Elife. 2015. PMID: 26258880 Free PMC article. - The PII-NAGK-PipX-NtcA Regulatory Axis of Cyanobacteria: A Tale of Changing Partners, Allosteric Effectors and Non-covalent Interactions.
Forcada-Nadal A, Llácer JL, Contreras A, Marco-Marín C, Rubio V. Forcada-Nadal A, et al. Front Mol Biosci. 2018 Nov 13;5:91. doi: 10.3389/fmolb.2018.00091. eCollection 2018. Front Mol Biosci. 2018. PMID: 30483512 Free PMC article. Review. - Recycling of Bacterial RNA Polymerase by the Swi2/Snf2 ATPase RapA.
Inlow K, Tenenbaum D, Friedman LJ, Kondev J, Gelles J. Inlow K, et al. bioRxiv [Preprint]. 2023 Mar 24:2023.03.22.533849. doi: 10.1101/2023.03.22.533849. bioRxiv. 2023. PMID: 36993374 Free PMC article. Updated. Preprint. - Crystal structure of the ATPase-C domain of the chromatin remodeller Fun30 from Saccharomyces cerevisiae.
Liu L, Jiang T. Liu L, et al. Acta Crystallogr F Struct Biol Commun. 2017 Jan 1;73(Pt 1):9-15. doi: 10.1107/S2053230X16019269. Epub 2017 Jan 1. Acta Crystallogr F Struct Biol Commun. 2017. PMID: 28045388 Free PMC article.
References
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. - PubMed
- Brown E, Malakar S, Krebs JE. How many remodelers does it take to make a brain? Diverse and cooperative roles of ATP-dependent chromatin-remodeling complexes in development. Biochem Cell Biol. 2007;85:444–462. - PubMed
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases