Hemiplasy and homoplasy in the karyotypic phylogenies of mammals - PubMed (original) (raw)
Hemiplasy and homoplasy in the karyotypic phylogenies of mammals
Terence J Robinson et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Phylogenetic reconstructions are often plagued by difficulties in distinguishing phylogenetic signal (due to shared ancestry) from phylogenetic noise or homoplasy (due to character-state convergences or reversals). We use a new interpretive hypothesis, termed hemiplasy, to show how random lineage sorting might account for specific instances of seeming "phylogenetic discordance" among different chromosomal traits, or between karyotypic features and probable species phylogenies. We posit that hemiplasy is generally less likely for underdominant chromosomal polymorphisms (i.e., those with heterozygous disadvantage) than for neutral polymorphisms or especially for overdominant rearrangements (which should tend to be longer-lived), and we illustrate this concept by using examples from chiropterans and afrotherians. Chromosomal states are especially powerful in phylogenetic reconstructions because they offer strong signatures of common ancestry, but their evolutionary interpretations remain fully subject to the principles of cladistics and the potential complications of hemiplasy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
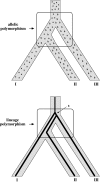
Fig. 1.
Schematic representations of hemiplasy. Shown are the distributions of a genic or chromosomal polymorphism (Upper) and a set of genealogical lineages (Lower) that traversed successive speciation nodes in an organismal phylogeny (broad branches) only to become fixed, by lineage sorting, in descendant species in a pattern that appears at face value to be discordant with the species phylogeny. In these diagrams, species II and III both have the homologous and derived character “b,” so the gene tree gives the impression that species II and III are the more closely related. However, in truth species I and II are sister taxa, despite the fact that species I alone retains the ancestral genetic condition “a.”
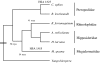
Fig. 2.
Phylogenetic relationships among bat species representing four taxonomic families (redrawn from ref. 27). Also shown is the presence of the chromosomal synteny 1/6/5, which formerly was interpreted to be homoplasic (27) but might instead be an example of hemiplasy.
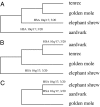
Fig. 3.
Competing phylogenetic hypotheses for species comprising the Afroinsectiphillia clade (aardvark, elephant shrew, golden mole, and tenrec). (A) Clade comprising the elephant shrew, tenrec, and golden mole to the exclusion of aardvark based on concatenations of nuclear and mitochondrial DNA sequences (40, 42). (B) Clade composed of aardvark plus tenrec and a weaker grouping of these taxa and the golden mole to the exclusion of elephant shrew based on a different concatenation (41). (C) Clade composed of aardvark and elephant shrew to the exclusion of golden mole based on cross-species CP (43).
Similar articles
- Examination of hemiplasy, homoplasy and phylogenetic discordance in chromosomal evolution of the Bovidae.
Robinson TJ, Ropiquet A. Robinson TJ, et al. Syst Biol. 2011 Jul;60(4):439-50. doi: 10.1093/sysbio/syr045. Epub 2011 Apr 28. Syst Biol. 2011. PMID: 21527771 - Phylotranscriptomic discordance is best explained by incomplete lineage sorting within Allium subgenus Cyathophora and thus hemiplasy accounts for interspecific trait transition.
Zhang Z, Liu G, Li M. Zhang Z, et al. Plant Divers. 2023 Jul 17;46(1):28-38. doi: 10.1016/j.pld.2023.07.004. eCollection 2024 Jan. Plant Divers. 2023. PMID: 38343588 Free PMC article. - Quantifying the risk of hemiplasy in phylogenetic inference.
Guerrero RF, Hahn MW. Guerrero RF, et al. Proc Natl Acad Sci U S A. 2018 Dec 11;115(50):12787-12792. doi: 10.1073/pnas.1811268115. Epub 2018 Nov 27. Proc Natl Acad Sci U S A. 2018. PMID: 30482861 Free PMC article. - Summary of Laurasiatheria (mammalia) phylogeny.
Hu JY, Zhang YP, Yu L. Hu JY, et al. Dongwuxue Yanjiu. 2012 Dec;33(E5-6):E65-74. doi: 10.3724/SP.J.1141.2012.E05-06E65. Dongwuxue Yanjiu. 2012. PMID: 23266984 Review.
Cited by
- Convergence of Afrotherian and Laurasiatherian Ungulate-Like Mammals: First Morphological Evidence from the Paleocene of Morocco.
Gheerbrant E, Filippo A, Schmitt A. Gheerbrant E, et al. PLoS One. 2016 Jul 6;11(7):e0157556. doi: 10.1371/journal.pone.0157556. eCollection 2016. PLoS One. 2016. PMID: 27384169 Free PMC article. - Karyotypic relationships in Asiatic asses (kulan and kiang) as defined using horse chromosome arm-specific and region-specific probes.
Musilova P, Kubickova S, Horin P, Vodicka R, Rubes J. Musilova P, et al. Chromosome Res. 2009;17(6):783-90. doi: 10.1007/s10577-009-9069-3. Chromosome Res. 2009. PMID: 19731053 - Chromosomal evolution in Raphicerus antelope suggests divergent X chromosomes may drive speciation through females, rather than males, contrary to Haldane's rule.
Robinson TJ, Cernohorska H, Kubickova S, Vozdova M, Musilova P, Ruiz-Herrera A. Robinson TJ, et al. Sci Rep. 2021 Feb 4;11(1):3152. doi: 10.1038/s41598-021-82859-0. Sci Rep. 2021. PMID: 33542477 Free PMC article. - Analysis of the Robertsonian (1;29) fusion in Bovinae reveals a common mechanism: insights into its clinical occurrence and chromosomal evolution.
Escudeiro A, Adega F, Robinson TJ, Heslop-Harrison JS, Chaves R. Escudeiro A, et al. Chromosome Res. 2021 Dec;29(3-4):301-312. doi: 10.1007/s10577-021-09667-0. Epub 2021 Jul 31. Chromosome Res. 2021. PMID: 34331632 - Extensive Chromosomal Reorganization in the Evolution of New World Muroid Rodents (Cricetidae, Sigmodontinae): Searching for Ancestral Phylogenetic Traits.
Pereira AL, Malcher SM, Nagamachi CY, O'Brien PC, Ferguson-Smith MA, Mendes-Oliveira AC, Pieczarka JC. Pereira AL, et al. PLoS One. 2016 Jan 22;11(1):e0146179. doi: 10.1371/journal.pone.0146179. eCollection 2016. PLoS One. 2016. PMID: 26800516 Free PMC article.
References
- Hennig W. Grundzüge einer Theorie der Phylogenetischen Systematik. Berlin: Deutscher Zentralverlag; 1950. (1996 Phylogenetic Systematics (University of Illinois Press, Urbana, Illinois) (English)
- Ferguson-Smith MA, Trifonov V. Mammalian karyotype evolution. Nat Rev Genet. 2007;8:950–962. - PubMed
- Avise JC, Robinson TJ. Hemiplasy: A new term in the lexicon of phylogenetics. Syst Biol. 2008;57:503–507. - PubMed
- Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ Press; 1987.
- Rosenberg NA. The probability of topological concordance of gene trees and species trees. Theor Pop Biol. 2002;61:225–247. - PubMed