Species-specific transcription in mice carrying human chromosome 21 - PubMed (original) (raw)
Species-specific transcription in mice carrying human chromosome 21
Michael D Wilson et al. Science. 2008.
Abstract
Homologous sets of transcription factors direct conserved tissue-specific gene expression, yet transcription factor-binding events diverge rapidly between closely related species. We used hepatocytes from an aneuploid mouse strain carrying human chromosome 21 to determine, on a chromosomal scale, whether interspecies differences in transcriptional regulation are primarily directed by human genetic sequence or mouse nuclear environment. Virtually all transcription factor-binding locations, landmarks of transcription initiation, and the resulting gene expression observed in human hepatocytes were recapitulated across the entire human chromosome 21 in the mouse hepatocyte nucleus. Thus, in homologous tissues, genetic sequence is largely responsible for directing transcriptional programs; interspecies differences in epigenetic machinery, cellular environment, and transcription factors themselves play secondary roles.
Figures
Figure 1
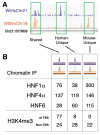
Transcriptional regulation varies between human and mouse hepatocytes across a complete chromosome. (A) Genome track showing chromatin immunoprecipitation enrichment of HNF1α binding in wild-type mouse and human hepatocytes across 30 kb of genomic sequence. The species of bound DNA sequences and ChIP signal is indicated by color: purple represents human, orange represents mouse. Highlighted in green are HNF1α-bound regions that are shared by both species, human-unique, and mouse-unique. (B) The total number of genomic regions occupied by three transcription factors (HNF1α, HNF4α, and HNF6) and the trimethylated form of histone H3K4 (H3K4me3) that are shared between the species, human-unique, or mouse-unique. ChIP data was obtained in wild-type mouse and human hepatocytes across the homologous regions of human chromosome 21 and mouse chromosome 16.
Figure 2
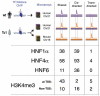
Comparison of the binding of the liver-specific transcription factors HNF1α, HNF4α, and HNF6, and enrichment of H3K4me3 on TcHsChr21 with the corresponding data obtained in mouse TcMmChr16 and human WtHsChr21 regions. The color scheme is the same as in Fig 1; notably, the primary difference from Fig 1 is the addition of the human chromosome in a mouse environment, which is indicated as a purple bar (representing the human chromosomal sequences) with an orange peak (from mouse transcription factor binding). The binding events on TcHsChr21 are sorted into categories based on whether they align with similar peaks in mouse and human (shared), align only with peaks in human (_cis_-directed), or align only with peaks in mice (_trans_-directed).
Figure 3
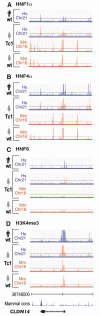
Patterns of transcription factor binding and transcription initiation are determined by genetic sequence. ChIP enrichment for (A) HNF1α, (B) HNF4α, (C) HNF6, and (D) H3K4me3 are shown across a 50 kb surrounding the liver-expressed gene CLDN14. The human chromosome 21 coordinates, and the vertebrate sequence conservation track (Seq Cons;
genome.ucsc.edu
) are shown flanking CLDN14. Each panel shows the species of genetic sequence as a bar colored by species (human: purple, mouse: orange) below a track showing ChIP enrichment, similarly colored by species.
Figure 4
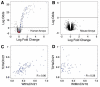
Gene expression in the Tc1 mouse originating from the mouse and human chromosomes is largely indistinguishable from comparable wild-type nuclear environments. Volcano plots (empirical Bayes log odds of differential expression versus average log fold change) demonstrating that (A) Tc1 hepatocytes have high transcription occurring from the transplanted human chromosome 21 using human genomic arrays and wild-type littermate mRNA as a reference (black probes map to human genes; blue probes map to genes located on HsChr21; red probes map to regions absent from TcHsChr21); whereas (B) wild-type and Tc1 mouse gene expression on mouse genomic arrays have indistinguishable patterns of transcription (black probes map to mouse genes). (C) Plot of the log expression of TcHsChr21 (y-axis) transcripts versus WtHsChr21 (x-axis) transcripts (Rcorr ≈ 0.90). (D) Plot of the log expression of TcHsChr21 (y-axis) transcripts versus WtMmChr16 (x-axis) orthologous transcripts (Rcorr ≈ 0.28).
Comment in
- Genetics. It's the sequence, stupid!
Coller HA, Kruglyak L. Coller HA, et al. Science. 2008 Oct 17;322(5900):380-1. doi: 10.1126/science.1165664. Science. 2008. PMID: 18927376 Free PMC article. No abstract available.
Similar articles
- Genetics. It's the sequence, stupid!
Coller HA, Kruglyak L. Coller HA, et al. Science. 2008 Oct 17;322(5900):380-1. doi: 10.1126/science.1165664. Science. 2008. PMID: 18927376 Free PMC article. No abstract available. - Mouse models of Down syndrome: how useful can they be? Comparison of the gene content of human chromosome 21 with orthologous mouse genomic regions.
Gardiner K, Fortna A, Bechtel L, Davisson MT. Gardiner K, et al. Gene. 2003 Oct 30;318:137-47. doi: 10.1016/s0378-1119(03)00769-8. Gene. 2003. PMID: 14585506 - Latent regulatory potential of human-specific repetitive elements.
Ward MC, Wilson MD, Barbosa-Morais NL, Schmidt D, Stark R, Pan Q, Schwalie PC, Menon S, Lukk M, Watt S, Thybert D, Kutter C, Kirschner K, Flicek P, Blencowe BJ, Odom DT. Ward MC, et al. Mol Cell. 2013 Jan 24;49(2):262-72. doi: 10.1016/j.molcel.2012.11.013. Epub 2012 Dec 13. Mol Cell. 2013. PMID: 23246434 Free PMC article. - The sequence of human chromosome 21 and implications for research into Down syndrome.
Gardiner K, Davisson M. Gardiner K, et al. Genome Biol. 2000;1(2):REVIEWS0002. doi: 10.1186/gb-2000-1-2-reviews0002. Epub 2000 Aug 4. Genome Biol. 2000. PMID: 11178230 Free PMC article. Review. - The transcriptional regulatory code of eukaryotic cells--insights from genome-wide analysis of chromatin organization and transcription factor binding.
Barrera LO, Ren B. Barrera LO, et al. Curr Opin Cell Biol. 2006 Jun;18(3):291-8. doi: 10.1016/j.ceb.2006.04.002. Epub 2006 May 2. Curr Opin Cell Biol. 2006. PMID: 16647254 Review.
Cited by
- A novel test for selection on cis-regulatory elements reveals positive and negative selection acting on mammalian transcriptional enhancers.
Smith JD, McManus KF, Fraser HB. Smith JD, et al. Mol Biol Evol. 2013 Nov;30(11):2509-18. doi: 10.1093/molbev/mst134. Epub 2013 Jul 30. Mol Biol Evol. 2013. PMID: 23904330 Free PMC article. - Improved predictions of transcription factor binding sites using physicochemical features of DNA.
Maienschein-Cline M, Dinner AR, Hlavacek WS, Mu F. Maienschein-Cline M, et al. Nucleic Acids Res. 2012 Dec;40(22):e175. doi: 10.1093/nar/gks771. Epub 2012 Aug 25. Nucleic Acids Res. 2012. PMID: 22923524 Free PMC article. - Comparative epigenomic annotation of regulatory DNA.
Xiao S, Xie D, Cao X, Yu P, Xing X, Chen CC, Musselman M, Xie M, West FD, Lewin HA, Wang T, Zhong S. Xiao S, et al. Cell. 2012 Jun 8;149(6):1381-92. doi: 10.1016/j.cell.2012.04.029. Cell. 2012. PMID: 22682255 Free PMC article. - Evidence that localized variation in primate sequence divergence arises from an influence of nucleosome placement on DNA repair.
Ying H, Epps J, Williams R, Huttley G. Ying H, et al. Mol Biol Evol. 2010 Mar;27(3):637-49. doi: 10.1093/molbev/msp253. Epub 2009 Oct 20. Mol Biol Evol. 2010. PMID: 19843619 Free PMC article. - Coevolution within a transcriptional network by compensatory trans and cis mutations.
Kuo D, Licon K, Bandyopadhyay S, Chuang R, Luo C, Catalana J, Ravasi T, Tan K, Ideker T. Kuo D, et al. Genome Res. 2010 Dec;20(12):1672-8. doi: 10.1101/gr.111765.110. Epub 2010 Oct 26. Genome Res. 2010. PMID: 20978140 Free PMC article.
References
- Davidson EH, Erwin DH. Science. 2006 Feb 10;311:796. - PubMed
- Zaret KS. Mech Dev. 2000 Mar 15;92:83. - PubMed
- Wray GA. Nat Rev Genet. 2007 Mar;8:206. - PubMed
- Li B, Carey M, Workman JL. Cell. 2007 Feb 23;128:707. - PubMed
- Guccione E, et al. Nat Cell Biol. 2006 Jul;8:764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 080174/WT_/Wellcome Trust/United Kingdom
- MC_U117527252/MRC_/Medical Research Council/United Kingdom
- 15603/CRUK_/Cancer Research UK/United Kingdom
- A15603/CRUK_/Cancer Research UK/United Kingdom
- G0601056/MRC_/Medical Research Council/United Kingdom
- 202218/ERC_/European Research Council/International
LinkOut - more resources
Full Text Sources
Other Literature Sources