The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure - PubMed (original) (raw)
. 2008 Sep 12;4(9):e1000190.
doi: 10.1371/journal.pgen.1000190.
Hui Su, Audesh Bhat, Hong Lei, Jeffrey Bajko, Sarah Hevi, Gretchen A Baltus, Shilpa Kadam, Huili Zhai, Reginald Valdez, Susana Gonzalo, Yi Zhang, En Li, Taiping Chen
Affiliations
- PMID: 18787701
- PMCID: PMC2527135
- DOI: 10.1371/journal.pgen.1000190
The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure
Brendan Jones et al. PLoS Genet. 2008.
Abstract
Dot1 is an evolutionarily conserved histone methyltransferase specific for lysine 79 of histone H3 (H3K79). In Saccharomyces cerevisiae, Dot1-mediated H3K79 methylation is associated with telomere silencing, meiotic checkpoint control, and DNA damage response. The biological function of H3K79 methylation in mammals, however, remains poorly understood. Using gene targeting, we generated mice deficient for Dot1L, the murine Dot1 homologue. Dot1L-deficient embryos show multiple developmental abnormalities, including growth impairment, angiogenesis defects in the yolk sac, and cardiac dilation, and die between 9.5 and 10.5 days post coitum. To gain insights into the cellular function of Dot1L, we derived embryonic stem (ES) cells from Dot1L mutant blastocysts. Dot1L-deficient ES cells show global loss of H3K79 methylation as well as reduced levels of heterochromatic marks (H3K9 di-methylation and H4K20 tri-methylation) at centromeres and telomeres. These changes are accompanied by aneuploidy, telomere elongation, and proliferation defects. Taken together, these results indicate that Dot1L and H3K79 methylation play important roles in heterochromatin formation and in embryonic development.
Conflict of interest statement
All authors except AB, SG, and YZ are employees of Novartis Institutes for Biomedical Research.
Figures
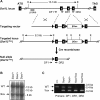
Figure 1. Generation of mutant Dot1L alleles in mice.
(A) Schematic depiction of the strategy used to generate the Dot1L3lox and Dot1L1lox alleles. The exons are numbered. The locations of the Southern probe and PCR primers (DF1, DR1, and DR2) used for genotyping, as well as the sizes of the diagnostic fragments recognized by the Southern probe, are indicated (E, _Eco_RI). loxP sites are shown as triangles. (B) Southern blot analysis of _Eco_RI-digested genomic DNA probed with an 860-bp 5′ probe external to the targeting vector. The presence of the 8.3-kb band confirms homologous recombination. (C) PCR genotyping of DNA from ES cells. WT, 485 bp; 1lox, 233 bp.
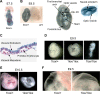
Figure 2. Essential role for Dot1L in mouse embryonic development.
(A) A representative X-gal stained 7.5-dpc Dot1L3lox/+ embryo demonstrating ubiquitous Dot1L transcription throughout the embryo. (B) A representative X-gal stained 9.5-dpc Dot1L3lox/+ embryo demonstrating ubiquitous Dot1L transcription throughout the embryo with elevated Dot1L expression in the indicated regions. (C) A representative X-gal stained 9.5-dpc Dot1L3lox/+ yolk sac demonstrating Dot1L transcription in visceral endoderm, visceral mesoderm, and primitive erythrocytes. (D) Representative pictures of 9.5-dpc Dot1L1lox/+ and Dot1L1lox/1lox embryos. Dot1L1lox/+ embryos (left) were indistinguishable from wild-type embryos. Most Dot1L1lox/1lox embryos were undersized, had an enlarged heart (cardiac dilation) and stunted tail (center), while approximately 15% exhibited developmental arrest at E8.5 (right). (E) Representative pictures of a 10.5-dpc Dot1L1lox/1lox embryo (right) and a heterozygous littermate (left). (F) Representative pictures showing the yolk sac vasculature of 9.5-dpc Dot1L1lox/+ (left) and Dot1L1lox/1lox (right) embryos. The vasculature of the Dot1L1lox/1lox yolk sac is thinner and less organized than that of the heterozygous littermate.
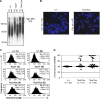
Figure 3. Phenotypic analysis of Dot1L mutant ES cells.
(A) Western blot analysis using extracts from ES cell lines of the indicated Dot1L genotypes and antibodies specific for di-, and tri-methylated H3K79. Total histone H3 was used as a loading control. (B) Analysis of H3K79 methylation by mass spectrometry. Quantification of different forms of H3K79 methylation was obtained by comparing the extracted ion chromatogram (EIC) intensity of the ion signals corresponding to the unmodified (Me0), mono-methylated (Me1), di-methylated (Me2), and tri-methylated (Me3) K79-containing peptides. (C) The proliferation of Dot1L+/+, Dot1L1lox/+ and Dot1L1lox/1lox ES cells was determined by doing cell counts every 24 hours for five days. Cells were grown in triplicate, and data shown is representative of three independent experiments. (D) The percentages of apoptotic cells in Dot1L+/+, Dot1L1lox/+ and Dot1L1lox/1lox ES cell cultures. The asterisk indicates P<0.05 (Student t-test). ES cells were stained with propidium iodide (PI) and PE conjugated anti-annexin V antibodies and analyzed by FACS. Apoptotic cells were annexin V positive and PI negative. Cells were grown in triplicate, and data shown are representative of two independent experiments. (E) The percentages of each cell cycle stage in Dot1L+/+, Dot1L1lox/+ and Dot1L1lox/1lox ES cell cultures as determined by PI staining and FACS.
Figure 4. Telomere elongation and aneuploidy in _Dot1L_-deficient ES cells.
(A) Telomere restriction fragment (TRF) analysis upon _Mbo_I digestion of genomic DNA from two independent ES cell clones of each of the genotypes: Dot1L+/+, Dot1L1lox/+ and Dot1L1lox/1lox. Note the presence of high molecular weight TRFs in Dot1L1lox/1lox cells, which correspond to longer telomeres. (B) Representative images generated during the Q-FISH assay showing metaphase spreads from Dot1L+/+ and Dot1L1lox/1lox ES cells labeled with a telomere-specific fluorescent probe. (C) Telomere length distribution of two independent ES cell clones of each of the Dot1L genotypes as determined by Q-FISH. Twenty metaphases of each ES cell clone were analyzed. Note the increase in mean telomere length (Mtl) in both clones of Dot11lox/1lox cells and the intermediate phenotype of Dot11lox/+ lines. The percentages of telomeres below 50 kb and above 100 kb in length are indicated. (D) Scatter plot of the chromosome number of a Dot1L+/+ ES cell line and two Dot1L1lox/1lox cell lines. Chromosome number was determined by manually counting chromosomes in chromosome spreads. Each point represents the chromosome number of a single cell (n represents the number of metaphase cells counted).
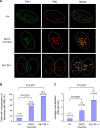
Figure 5. Increased APBs in _Dot1L_-deficient cells.
(A) Confocal microscopy images showing either TRF1 (telomere marker, green), PML (marker for PML bodies, red), or combined fluorescence (yellow if colocalize, indicated by arrows) in wild-type (+/+) and _Dot1L_-deficient (1lox/1lox) ES cells. Late-passage (p120) _Dnmt3a/3b_-deficient (3a−/−3b−/−) ES cells were used as a positive control. Circled are nuclei of cells. (B) Quantification of percentage of cells showing colocalization of telomeres with PML bodies. A cell was considered positive when it showed 2 or more colocalization events. An increased frequency of cells showing APBs was observed in Dot11lox/1lox cultures compared to wild-type controls (χ2 test, P<0.001). (C) Quantification of the number of APBs per cell. Dot11lox/1lox cells showed a significant increase in the number of APBs compared to wild-type cells (χ2 test, P<0.001).
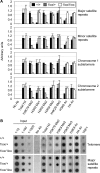
Figure 6. Changes of heterochromatin structure in _Dot1L_-deficient ES cells.
(A) Quantitative real-time PCR results using DNA from Dot1L mutant and wild-type ES cells immunoprecipitated with antibodies specific for the indicated histone modifications or without an antibody (No Ab) and normalized using input DNA values. PCR primers specific for major satellite repeats, minor satellite repeats, the subtelomere region of chromosome 1 or the subtelomere region of chromosome 2 were used. (B) Dot blot analysis of ChIP DNA using either a telomere-specific probe or a major satellite repeat-specific probe. Input DNA at 1∶10, 1∶100 and 1∶1000 dilutions was used as a positive control. DNA precipitated from 2.5×106 cells were used for each assay.
Similar articles
- The histone methyltransferase Dot1/DOT1L as a critical regulator of the cell cycle.
Kim W, Choi M, Kim JE. Kim W, et al. Cell Cycle. 2014;13(5):726-38. doi: 10.4161/cc.28104. Epub 2014 Feb 6. Cell Cycle. 2014. PMID: 24526115 Free PMC article. Review. - Deficiency of H3K79 histone methyltransferase Dot1-like protein (DOT1L) inhibits cell proliferation.
Kim W, Kim R, Park G, Park JW, Kim JE. Kim W, et al. J Biol Chem. 2012 Feb 17;287(8):5588-99. doi: 10.1074/jbc.M111.328138. Epub 2011 Dec 21. J Biol Chem. 2012. PMID: 22190683 Free PMC article. - Dot1L mediated histone H3 lysine79 methylation is essential to meiosis progression in mouse oocytes.
Wang X, Gao W, Ma X, Wang X, Song C, Huang X, Liu H. Wang X, et al. Neuro Endocrinol Lett. 2014;35(6):523-30. Neuro Endocrinol Lett. 2014. PMID: 25433842 - DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells.
Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, Blobel GA, Vakoc CR. Steger DJ, et al. Mol Cell Biol. 2008 Apr;28(8):2825-39. doi: 10.1128/MCB.02076-07. Epub 2008 Feb 19. Mol Cell Biol. 2008. PMID: 18285465 Free PMC article. - The diverse functions of Dot1 and H3K79 methylation.
Nguyen AT, Zhang Y. Nguyen AT, et al. Genes Dev. 2011 Jul 1;25(13):1345-58. doi: 10.1101/gad.2057811. Genes Dev. 2011. PMID: 21724828 Free PMC article. Review.
Cited by
- Histone H3K79 methyltransferase Dot1L is directly activated by thyroid hormone receptor during Xenopus metamorphosis.
Matsuura K, Fujimoto K, Das B, Fu L, Lu CD, Shi YB. Matsuura K, et al. Cell Biosci. 2012 Jul 16;2(1):25. doi: 10.1186/2045-3701-2-25. Cell Biosci. 2012. PMID: 22800560 Free PMC article. - Clock genes and their genomic distributions in three species of salmonid fishes: Associations with genes regulating sexual maturation and cell cycling.
Paibomesai MI, Moghadam HK, Ferguson MM, Danzmann RG. Paibomesai MI, et al. BMC Res Notes. 2010 Jul 29;3:215. doi: 10.1186/1756-0500-3-215. BMC Res Notes. 2010. PMID: 20670436 Free PMC article. - Histone Methyltransferase DOT1L is Involved in Larval Molting and Second Stage Nymphal Feeding in Ornithodoros Moubata.
Gobl J, Kumar Sinha D, Sima R, Perner J, Kopáček P, Valdés JJ, Rego ROM, Cabezas-Cruz A. Gobl J, et al. Vaccines (Basel). 2020 Apr 1;8(2):157. doi: 10.3390/vaccines8020157. Vaccines (Basel). 2020. PMID: 32244625 Free PMC article. - Disruptor of telomeric silencing 1-like (DOT1L): disclosing a new class of non-nucleoside inhibitors by means of ligand-based and structure-based approaches.
Sabatino M, Rotili D, Patsilinakos A, Forgione M, Tomaselli D, Alby F, Arimondo PB, Mai A, Ragno R. Sabatino M, et al. J Comput Aided Mol Des. 2018 Mar;32(3):435-458. doi: 10.1007/s10822-018-0096-z. Epub 2018 Jan 15. J Comput Aided Mol Des. 2018. PMID: 29335872 - The protective role of DOT1L in UV-induced melanomagenesis.
Zhu B, Chen S, Wang H, Yin C, Han C, Peng C, Liu Z, Wan L, Zhang X, Zhang J, Lian CG, Ma P, Xu ZX, Prince S, Wang T, Gao X, Shi Y, Liu D, Liu M, Wei W, Wei Z, Pan J, Wang Y, Xuan Z, Hess J, Hayward NK, Goding CR, Chen X, Zhou J, Cui R. Zhu B, et al. Nat Commun. 2018 Jan 17;9(1):259. doi: 10.1038/s41467-017-02687-7. Nat Commun. 2018. PMID: 29343685 Free PMC article.
References
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. - PubMed
- Janzen CJ, Hake SB, Lowell JE, Cross GA. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol Cell. 2006;23:497–507. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases